

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125478.4 + phase: 0
(260 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
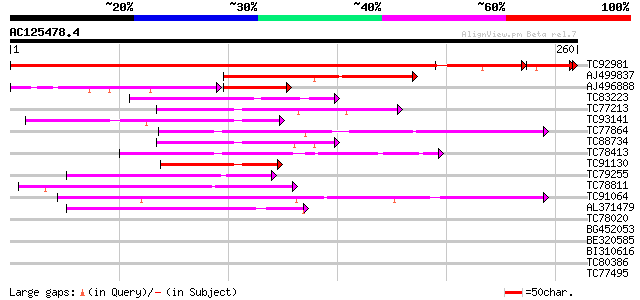
TC92981 similar to PIR|T47758|T47758 hypothetical protein F24I3.... 406 e-121
AJ499837 similar to PIR|T47758|T477 hypothetical protein F24I3.6... 106 8e-24
AJ496888 similar to PIR|T47757|T477 hypothetical protein F24I3.5... 62 3e-10
TC83223 similar to GP|8777436|dbj|BAA97026.1 gb|AAC78547.1~gene_... 52 2e-07
TC77213 similar to PIR|T10861|T10861 phaseolin G-box binding pro... 51 4e-07
TC93141 similar to GP|18491261|gb|AAL69455.1 At2g41130/T3K9.10 {... 47 6e-06
TC77864 weakly similar to PIR|T06329|T06329 symbiotic ammonium t... 46 2e-05
TC88734 similar to GP|13346180|gb|AAK19612.1 GHDEL61 {Gossypium ... 44 6e-05
TC78413 similar to PIR|T06329|T06329 symbiotic ammonium transpor... 44 8e-05
TC91130 homologue to GP|11245494|gb|AAG33640.1 SPATULA {Arabidop... 43 1e-04
TC79255 similar to PIR|E96714|E96714 probable DNA-binding protei... 43 1e-04
TC78811 similar to GP|3641870|emb|CAA09459.1 hypothetical protei... 43 1e-04
TC91064 weakly similar to GP|20127014|gb|AAM10934.1 putative bHL... 42 2e-04
AL371479 similar to PIR|E96714|E967 probable DNA-binding protein... 41 4e-04
TC78020 similar to GP|20127060|gb|AAM10949.1 putative bHLH trans... 40 0.001
BG452053 homologue to GP|11994233|db contains similarity to heli... 40 0.001
BE320585 similar to GP|10177877|dbj gene_id:MIO24.9~pir||T10220~... 39 0.002
BI310616 similar to GP|21593074|gb| bHLH DNA-binding protein-lik... 39 0.002
TC80386 weakly similar to GP|10800070|dbj|BAB16490. contains EST... 39 0.002
TC77495 similar to GP|2673918|gb|AAB88652.1| unknown protein {Ar... 39 0.002
>TC92981 similar to PIR|T47758|T47758 hypothetical protein F24I3.60 -
Arabidopsis thaliana, partial (27%)
Length = 828
Score = 406 bits (1043), Expect(2) = e-121
Identities = 215/241 (89%), Positives = 218/241 (90%), Gaps = 4/241 (1%)
Frame = +3
Query: 1 MVAFCPPQFSYSNMGWLLEELEPESLISHKEKNYASLEYSLPYHQFSSPKEHVEIERPPS 60
MVAFCPPQFSYSNMGWLLEELEPESLISHKEKNYASLEYSLPYHQFSSPKEHVEIERPPS
Sbjct: 21 MVAFCPPQFSYSNMGWLLEELEPESLISHKEKNYASLEYSLPYHQFSSPKEHVEIERPPS 200
Query: 61 PKLMAKKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQ 120
PKLMAKKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQ
Sbjct: 201 PKLMAKKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQ 380
Query: 121 VQGLTKKKEELLSRISHRQEYAVNKESQRKKIPNYNSAFVVSTSRLNDTELVIHISSYEA 180
VQGLTKKKEELLSRISHRQEYAVNKESQRKKIPNYNSAFVVSTSRLNDTELVIHISSYEA
Sbjct: 381 VQGLTKKKEELLSRISHRQEYAVNKESQRKKIPNYNSAFVVSTSRLNDTELVIHISSYEA 560
Query: 181 NKIPLSEILMCLENNGLLLLNSSSSKTFGGRLFYN----LHFQVDKTQRYECDDLIQKLS 236
NKIPLSEILMCLENNG +SS+ F L+ L F RYECDDLIQKLS
Sbjct: 561 NKIPLSEILMCLENNG----SSST*LFFL*NLWREALL*LAFSGG*NSRYECDDLIQKLS 728
Query: 237 S 237
S
Sbjct: 729 S 731
Score = 62.4 bits (150), Expect = 2e-10
Identities = 38/66 (57%), Positives = 42/66 (63%), Gaps = 2/66 (3%)
Frame = +1
Query: 196 GLLLLNSSSSKTFGGRLFYNLHFQVDKTQRYECDDLIQKLSSIYE--KQQNNHLGTMDQT 253
GLLLLNSSSSKTFGGRLFYNLHFQVDKTQ + I E K H G+ DQ
Sbjct: 607 GLLLLNSSSSKTFGGRLFYNLHFQVDKTQDMSVMI*FKSFLHI*EAAK*SFGHYGSNDQ* 786
Query: 254 INSVLI 259
+ +LI
Sbjct: 787 WSDILI 804
Score = 48.1 bits (113), Expect(2) = e-121
Identities = 22/23 (95%), Positives = 22/23 (95%)
Frame = +2
Query: 238 IYEKQQNNHLGTMDQTINSVLIY 260
IYEKQQNNHLGTMDQTINS LIY
Sbjct: 731 IYEKQQNNHLGTMDQTINSGLIY 799
>AJ499837 similar to PIR|T47758|T477 hypothetical protein F24I3.60 -
Arabidopsis thaliana, partial (13%)
Length = 484
Score = 106 bits (265), Expect = 8e-24
Identities = 57/90 (63%), Positives = 73/90 (80%), Gaps = 1/90 (1%)
Frame = +3
Query: 99 KKMSIPVTISRVLKYIPDLQKQVQGLTKKKEELLSRISHR-QEYAVNKESQRKKIPNYNS 157
KK+SIP TISRVLKYIP+LQ QV+GL K+K+E+L +S + +E+ ++KESQRKK +YNS
Sbjct: 216 KKLSIPGTISRVLKYIPELQNQVEGLIKRKDEILLGLSPQVEEFILSKESQRKK-HSYNS 392
Query: 158 AFVVSTSRLNDTELVIHISSYEANKIPLSE 187
FVVS+SRLND+E+ I IS Y KIPLSE
Sbjct: 393 GFVVSSSRLNDSEITIQISCYTVQKIPLSE 482
>AJ496888 similar to PIR|T47757|T477 hypothetical protein F24I3.50 -
Arabidopsis thaliana, partial (13%)
Length = 676
Score = 61.6 bits (148), Expect = 3e-10
Identities = 45/100 (45%), Positives = 56/100 (56%), Gaps = 3/100 (3%)
Frame = -3
Query: 1 MVAFCPPQFSYSNMGWLLEELEPESLISHKEKNYA-SLEYSLPYH-QFSSPKEHVEIERP 58
M+A PP FS +GW EE P S H+ Y +++ +H Q + + +
Sbjct: 539 MLAISPPMFS--TIGWPFEE--PLSHNQHQNSFYKDTVDQLFNFHDQVEAEINSTDPSQS 372
Query: 59 PSPKL-MAKKLNHNASERDRRKKINSLISSLRSLLPGEDQ 97
S L M KKL H ASERDRRKKIN+L SSLRSLLP DQ
Sbjct: 371 TSSDLSMVKKLVHYASERDRRKKINNLYSSLRSLLPVSDQ 252
Score = 51.2 bits (121), Expect = 4e-07
Identities = 24/31 (77%), Positives = 28/31 (89%)
Frame = +3
Query: 99 KKMSIPVTISRVLKYIPDLQKQVQGLTKKKE 129
KK+SIP TISRVLKYIP+LQ QV+GL K+KE
Sbjct: 144 KKLSIPGTISRVLKYIPELQNQVEGLIKRKE 236
>TC83223 similar to GP|8777436|dbj|BAA97026.1
gb|AAC78547.1~gene_id:MHM17.7~similar to unknown protein
{Arabidopsis thaliana}, partial (13%)
Length = 514
Score = 52.4 bits (124), Expect = 2e-07
Identities = 33/96 (34%), Positives = 54/96 (55%)
Frame = +1
Query: 56 ERPPSPKLMAKKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIP 115
+R + + + +L+H SER RR+K+N +LR+LLP + K SI +T L+
Sbjct: 160 DRNQAARPSSNQLHHMISERRRREKLNENFQALRALLPQGTKKDKASILITAKETLR--- 330
Query: 116 DLQKQVQGLTKKKEELLSRISHRQEYAVNKESQRKK 151
L ++ L+K+ +EL+S +Q A NKES + K
Sbjct: 331 SLMAEIDKLSKRNQELMS----QQLPAANKESTKTK 426
>TC77213 similar to PIR|T10861|T10861 phaseolin G-box binding protein PG1 -
kidney bean, partial (70%)
Length = 2391
Score = 51.2 bits (121), Expect = 4e-07
Identities = 34/120 (28%), Positives = 59/120 (48%), Gaps = 7/120 (5%)
Frame = +3
Query: 68 LNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKK 127
LNH +ER RR+K+N +LR+++P + K S+ + + YI +L+ ++QGL
Sbjct: 1542 LNHVEAERQRREKLNQKFYALRAVVPNVSKMDKASL---LGDAISYINELKSKLQGLESS 1712
Query: 128 KEEL---LSRISHRQEYAVNKESQRKKIP----NYNSAFVVSTSRLNDTELVIHISSYEA 180
K EL L E +K + IP + S+S+L D ++ + I ++A
Sbjct: 1713 KGELEKQLGATKKELELVASKNQSQNPIPLDKEKEKTTSSTSSSKLIDLDIDVKIMGWDA 1892
>TC93141 similar to GP|18491261|gb|AAL69455.1 At2g41130/T3K9.10 {Arabidopsis
thaliana}, partial (57%)
Length = 839
Score = 47.4 bits (111), Expect = 6e-06
Identities = 36/121 (29%), Positives = 62/121 (50%), Gaps = 2/121 (1%)
Frame = +2
Query: 8 QFSYSNMGWLLEELEPESLISHKEKNYASLEYSLPYHQFSSPKEHVEIERPPSP--KLMA 65
QF +N E P S + + +S + YH F E EI PS + +A
Sbjct: 227 QFLVANNPSFFEYSTPSSTMMQQSFCSSSDNNNNYYHPF----EVSEITDTPSQQDRALA 394
Query: 66 KKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLT 125
NH +E+ RR++INS + LR+LLP +T K S+ +++V++ + +L++Q +T
Sbjct: 395 ALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTDKASL---LAKVVERVKELKQQTSQIT 565
Query: 126 K 126
+
Sbjct: 566 Q 568
>TC77864 weakly similar to PIR|T06329|T06329 symbiotic ammonium transport
protein SAT1 - soybean, partial (21%)
Length = 1411
Score = 45.8 bits (107), Expect = 2e-05
Identities = 40/182 (21%), Positives = 82/182 (44%), Gaps = 3/182 (1%)
Frame = +3
Query: 69 NHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKKK 128
+H +ER RR+++ I +L +++PG KKM +S + Y LQK+++ L +
Sbjct: 558 DHLMAERKRRRELTENIIALSAMIPG---LKKMDKCYVLSEAVNYTKQLQKRIKELENQN 728
Query: 129 EELLSR---ISHRQEYAVNKESQRKKIPNYNSAFVVSTSRLNDTELVIHISSYEANKIPL 185
++ + + + NK+ + + + +R+ + E++I I + I L
Sbjct: 729 KDSKPNPAIFKWKSQVSSNKKKS-------SESLLEVEARVKEKEVLIRIHCEKQKDIVL 887
Query: 186 SEILMCLENNGLLLLNSSSSKTFGGRLFYNLHFQVDKTQRYECDDLIQKLSSIYEKQQNN 245
+I LE + + +SS L N+ Q+D+ DDL++ L + +
Sbjct: 888 -KIHELLEKFNITITSSSMLPFGDSILVINICAQMDEEDSMTMDDLVENLRKYLLETHES 1064
Query: 246 HL 247
+L
Sbjct: 1065YL 1070
>TC88734 similar to GP|13346180|gb|AAK19612.1 GHDEL61 {Gossypium hirsutum},
partial (40%)
Length = 1701
Score = 43.9 bits (102), Expect = 6e-05
Identities = 30/95 (31%), Positives = 49/95 (51%), Gaps = 11/95 (11%)
Frame = +3
Query: 68 LNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKK 127
+NH SER RR K+N +LRS++P + K+SI + ++Y+ L+K+++ L +
Sbjct: 1401 MNHVLSERRRRAKLNERFLTLRSMVPSNSKDDKVSI---LDDAIEYLRKLEKKIRELEAQ 1571
Query: 128 KE----ELLSRISHR-------QEYAVNKESQRKK 151
+E E S+ SH Y NK + KK
Sbjct: 1572 REPTGIESRSKKSHHDMVERTTDHYYNNKTNNGKK 1676
>TC78413 similar to PIR|T06329|T06329 symbiotic ammonium transport protein
SAT1 - soybean, partial (30%)
Length = 1397
Score = 43.5 bits (101), Expect = 8e-05
Identities = 39/149 (26%), Positives = 74/149 (49%)
Frame = +2
Query: 51 EHVEIERPPSPKLMAKKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRV 110
E ++ + + + +A +H +ER RR+K++ + +L +L+PG KKM +
Sbjct: 476 ETIKPQGQGTKRSVAHNQDHIIAERKRREKLSQCLIALAALIPG---LKKMDKASVLGDA 646
Query: 111 LKYIPDLQKQVQGLTKKKEELLSRISHRQEYAVNKESQRKKIPNYNSAFVVSTSRLNDTE 170
+KY+ +LQ++++ L ++ + SH Q V + Q+ + NS S N+T
Sbjct: 647 IKYVKELQERLRVLEEQNKN-----SHVQS-VVTVDEQQLSYDSSNSDDSEVASGNNET- 805
Query: 171 LVIHISSYEANKIPLSEILMCLENNGLLL 199
+ H+ + +K L I C + GLLL
Sbjct: 806 -LPHVEAKVLDKDVLIRI-HCQKQKGLLL 886
>TC91130 homologue to GP|11245494|gb|AAG33640.1 SPATULA {Arabidopsis
thaliana}, partial (20%)
Length = 766
Score = 43.1 bits (100), Expect = 1e-04
Identities = 21/56 (37%), Positives = 36/56 (63%)
Frame = +2
Query: 70 HNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLT 125
HN SE+ RR +IN + +L++L+P ++T K S+ + ++Y+ LQ QVQ L+
Sbjct: 44 HNLSEKRRRSRINEKMKALQNLIPNSNKTDKASM---LDEAIEYLKQLQLQVQMLS 202
>TC79255 similar to PIR|E96714|E96714 probable DNA-binding protein T6L1.19
[imported] - Arabidopsis thaliana, partial (30%)
Length = 847
Score = 43.1 bits (100), Expect = 1e-04
Identities = 27/96 (28%), Positives = 47/96 (48%)
Frame = +2
Query: 27 ISHKEKNYASLEYSLPYHQFSSPKEHVEIERPPSPKLMAKKLNHNASERDRRKKINSLIS 86
++H+E Y E++ Q +S + + + K + H+ +E+ RR KIN
Sbjct: 176 VNHEEDEYDDDEFNSNKKQATSSVPNTNKDGKATDKASVIRSKHSVTEQRRRSKINERFQ 355
Query: 87 SLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQ 122
LR L+P DQ K + V++Y+ LQ++VQ
Sbjct: 356 ILRDLIPQCDQ--KRDTASFLLEVIEYVQYLQERVQ 457
>TC78811 similar to GP|3641870|emb|CAA09459.1 hypothetical protein {Cicer
arietinum}, partial (90%)
Length = 1182
Score = 42.7 bits (99), Expect = 1e-04
Identities = 33/135 (24%), Positives = 59/135 (43%), Gaps = 7/135 (5%)
Frame = +2
Query: 5 CPPQFSYSNMG-------WLLEELEPESLISHKEKNYASLEYSLPYHQFSSPKEHVEIER 57
CPP ++N L E E + S N + S + PK R
Sbjct: 230 CPPYVDWNNNNSSTFSPPHLNEVQETTTDPSSNTNNTQNFHASPSVNTVIRPKRRRARSR 409
Query: 58 PPSPKLMAKKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDL 117
++ +++ H A ER+RRK++N +S LRSL+P + ++ I + ++ +L
Sbjct: 410 KNKEEIENQRMTHIAVERNRRKQMNEYLSILRSLMP-DSHIQRGDQASIIGGAINFVKEL 586
Query: 118 QKQVQGLTKKKEELL 132
+ + L KKE ++
Sbjct: 587 EHKFHFLGAKKERVV 631
>TC91064 weakly similar to GP|20127014|gb|AAM10934.1 putative bHLH
transcription factor {Arabidopsis thaliana}, partial
(19%)
Length = 751
Score = 42.4 bits (98), Expect = 2e-04
Identities = 49/246 (19%), Positives = 101/246 (40%), Gaps = 21/246 (8%)
Frame = +2
Query: 23 PESLISHKEKNYASLEYSLPYHQFSSPKEHVEIERPP-------------SPKLMAKKLN 69
P L+++ N + +P + S P + P P+ + L+
Sbjct: 2 PTDLVNNSSINVSQHAEEMPTNSLSIPTTEQHHDSLPLSSSTANQGSNSKKPRNTSDTLD 181
Query: 70 HNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKKKE 129
H SER+RR+ + S I L +L+PG + K+ + +K + + K+++ KKK+
Sbjct: 182 HIMSERNRRQLLTSKIIELSALIPGLKKIDKVHVVTEAINYMKQLEERLKELEEDIKKKD 361
Query: 130 E------LLSRISHRQEYAVNKESQRKKIPNYNSAFVVSTSRLNDTELVIHI--SSYEAN 181
SR+ ++ A+ E ++ N + + +R+ + E++I I E
Sbjct: 362 AGSLSTITRSRVLIDKDIAIG-EMNTEECYGRNESLLEVEARILEKEVLIKIYCGMQEGI 538
Query: 182 KIPLSEILMCLENNGLLLLNSSSSKTFGGRLFYNLHFQVDKTQRYECDDLIQKLSSIYEK 241
+ + L L L + S + FG L + ++ DL++KL +
Sbjct: 539 VVNIMSQLQLLH----LSITSINVLPFGNTLDITIIAKMGDKYNLTIKDLVKKLRVVATL 706
Query: 242 QQNNHL 247
Q ++++
Sbjct: 707 QVSHNV 724
>AL371479 similar to PIR|E96714|E967 probable DNA-binding protein T6L1.19
[imported] - Arabidopsis thaliana, partial (26%)
Length = 423
Score = 41.2 bits (95), Expect = 4e-04
Identities = 30/112 (26%), Positives = 53/112 (46%), Gaps = 1/112 (0%)
Frame = +1
Query: 27 ISHKEKNYASLEYSLPYHQFSSPKEHVEIERPPSPKLMAKKLNHNASERDRRKKINSLIS 86
++H+E Y E++ Q +S + + + K + H+ +E+ RR KIN
Sbjct: 100 VNHEEDEYDDDEFNSNKKQATSSVPNTNKDGKATDKASVIRSKHSVTEQRRRSKINERFQ 279
Query: 87 SLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKKKEELLS-RISH 137
LR L+P DQ + + + RV KY + QG +++ +L+ R SH
Sbjct: 280 ILRDLIPQCDQKRDTASFLLEERVQKY----EGSYQGWSQEPSKLMPWRNSH 423
>TC78020 similar to GP|20127060|gb|AAM10949.1 putative bHLH transcription
factor {Arabidopsis thaliana}, partial (30%)
Length = 1473
Score = 39.7 bits (91), Expect = 0.001
Identities = 25/87 (28%), Positives = 46/87 (52%)
Frame = +3
Query: 49 PKEHVEIERPPSPKLMAKKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTIS 108
P HV + R + +H+ +ER RR+KIN+ + L+ L+PG + K + +
Sbjct: 639 PYVHVRVRRGQATD------SHSLAERARREKINARMKLLQELVPGCE--KISGTALVLD 794
Query: 109 RVLKYIPDLQKQVQGLTKKKEELLSRI 135
++ ++ LQ+QV+ L+ K + RI
Sbjct: 795 EIINHVQTLQRQVEILSMKLAAVNPRI 875
>BG452053 homologue to GP|11994233|db contains similarity to helix-loop-helix
DNA-binding protein~gene_id:MUJ8.4 {Arabidopsis
thaliana}, partial (27%)
Length = 613
Score = 39.7 bits (91), Expect = 0.001
Identities = 23/81 (28%), Positives = 48/81 (58%), Gaps = 1/81 (1%)
Frame = +1
Query: 60 SPKLMAKKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQK 119
S ++ ++++ H A ER+RRK++N + LRSL+PG ++ I ++++ +L++
Sbjct: 274 SEEVESQRMTHIAVERNRRKQMNEHLRVLRSLMPG-SYVQRGDQASIIGGAIEFVRELEQ 450
Query: 120 QVQGL-TKKKEELLSRISHRQ 139
+Q L ++K+ LL +Q
Sbjct: 451 LLQCLXSQKRRRLLGEAQSKQ 513
>BE320585 similar to GP|10177877|dbj gene_id:MIO24.9~pir||T10220~similar to
unknown protein {Arabidopsis thaliana}, partial (33%)
Length = 491
Score = 39.3 bits (90), Expect = 0.002
Identities = 22/63 (34%), Positives = 38/63 (59%)
Frame = +2
Query: 66 KKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLT 125
KK+ H E+ RR+++ +L +SLRSLLP K S+ ++ + YI L+K ++ L+
Sbjct: 236 KKMVHREIEKQRRQEMATLHTSLRSLLPLHFIKGKRSLSDQMNEAVNYINHLKKNMKELS 415
Query: 126 KKK 128
K+
Sbjct: 416 YKR 424
>BI310616 similar to GP|21593074|gb| bHLH DNA-binding protein-like protein
{Arabidopsis thaliana}, partial (24%)
Length = 656
Score = 39.3 bits (90), Expect = 0.002
Identities = 20/76 (26%), Positives = 45/76 (58%), Gaps = 1/76 (1%)
Frame = +2
Query: 66 KKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLT 125
+++ H ER+RRK++N ++ LRSL+P E ++ + ++++ +L+ +Q L
Sbjct: 431 QRITHITVERNRRKQMNEHLAVLRSLMP-ESYVQRGDQASIVGGAIEFVKELEHLLQSLE 607
Query: 126 KKKEELLSR-ISHRQE 140
+K +L+ + ++H E
Sbjct: 608 ARKLQLVQQEVTHNNE 655
>TC80386 weakly similar to GP|10800070|dbj|BAB16490. contains ESTs
C61063(AU063369) C61063(AU093310)~unknown protein,
partial (17%)
Length = 655
Score = 39.3 bits (90), Expect = 0.002
Identities = 24/62 (38%), Positives = 39/62 (62%), Gaps = 2/62 (3%)
Frame = +1
Query: 53 VEIERPP-SPKLMAKKL-NHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRV 110
V++ER SP+ + L NH+ +ER RR +INS + +LRS++PG ++ K S+ +
Sbjct: 469 VKLERKGVSPERSIEALKNHSEAERRRRARINSHLDTLRSVIPGANKLDKASLLAEVITH 648
Query: 111 LK 112
LK
Sbjct: 649 LK 654
>TC77495 similar to GP|2673918|gb|AAB88652.1| unknown protein {Arabidopsis
thaliana}, partial (28%)
Length = 834
Score = 38.9 bits (89), Expect = 0.002
Identities = 29/90 (32%), Positives = 49/90 (54%), Gaps = 4/90 (4%)
Frame = +3
Query: 45 QFSSPKEHVEIERPPSPKLMAKK--LNHNAS--ERDRRKKINSLISSLRSLLPGEDQTKK 100
+ +S ++ ++++ K+ AK+ H S ER RR +I+ I L+ L P D K+
Sbjct: 255 KMTSMEKFLQVQGSVPCKIRAKRGFATHPRSIAERVRRTRISDRIKKLQGLFPKSD--KQ 428
Query: 101 MSIPVTISRVLKYIPDLQKQVQGLTKKKEE 130
S + ++YI DLQ+QVQ LT K++
Sbjct: 429 TSTADMLDLAVEYIKDLQEQVQILTDCKDK 518
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.131 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,396,579
Number of Sequences: 36976
Number of extensions: 120155
Number of successful extensions: 577
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 573
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 575
length of query: 260
length of database: 9,014,727
effective HSP length: 94
effective length of query: 166
effective length of database: 5,538,983
effective search space: 919471178
effective search space used: 919471178
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 57 (26.6 bits)
Medicago: description of AC125478.4


