

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125477.15 - phase: 0
(738 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
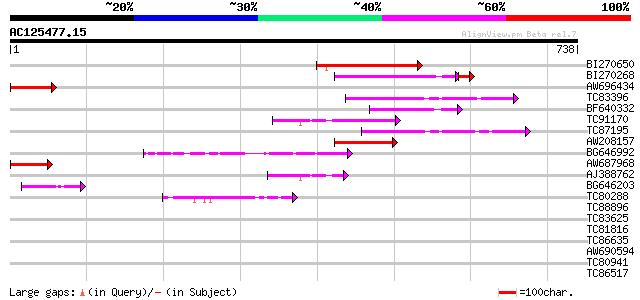
Score E
Sequences producing significant alignments: (bits) Value
BI270650 similar to PIR|T05016|T05 hypothetical protein T19P19.1... 152 3e-37
BI270268 weakly similar to GP|15724286|gb| AT4g35240/F23E12_200 ... 104 5e-24
AW696434 similar to GP|15810479|gb| unknown protein {Arabidopsis... 100 2e-21
TC83396 similar to GP|22858664|gb|AAN05792.1 unknown {Gossypium ... 92 9e-19
BF640332 weakly similar to GP|5263327|gb| EST gb|T20649 comes fr... 87 2e-17
TC91170 similar to PIR|D96563|D96563 probable bZIP protein 4865... 83 4e-16
TC87195 similar to GP|16604629|gb|AAL24107.1 putative bZIP prote... 72 6e-13
AW208157 similar to GP|16604629|gb putative bZIP protein {Arabid... 70 4e-12
BG646992 similar to PIR|T06133|T061 hypothetical protein F23E12.... 67 2e-11
AW687968 similar to GP|22136982|gb| unknown protein {Arabidopsis... 66 6e-11
AJ388762 homologue to PIR|D96563|D965 probable bZIP protein 486... 49 5e-06
BG646203 homologue to PIR|S22697|S226 extensin - Volvox carteri ... 46 6e-05
TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein... 43 5e-04
TC88896 weakly similar to GP|6728968|gb|AAF26966.1| unknown prot... 41 0.002
TC83625 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 40 0.004
TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 40 0.004
TC86635 weakly similar to GP|9758600|dbj|BAB09233.1 gene_id:MDF2... 37 0.027
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 37 0.027
TC80941 weakly similar to PIR|T39903|T39903 serine-rich protein ... 37 0.027
TC86517 homologue to PIR|T09640|T09640 protein phosphatase 2C - ... 34 0.23
>BI270650 similar to PIR|T05016|T05 hypothetical protein T19P19.180 -
Arabidopsis thaliana, partial (21%)
Length = 498
Score = 152 bits (385), Expect = 3e-37
Identities = 74/141 (52%), Positives = 102/141 (71%), Gaps = 3/141 (2%)
Frame = +1
Query: 400 SKTRDEVYDDPS---EEQCVFSVSHQSTLDRLYAWEKKLYQEVKSGTRVRLAYEKKCLQL 456
SK+R+++ D S EE C+ + SH STLDRLYAWE+KLY EVK+ +R Y++KC QL
Sbjct: 7 SKSREDIDDSGSNFVEEFCMIAGSHSSTLDRLYAWERKLYDEVKASESIRKVYDRKCHQL 186
Query: 457 RNHDIKGEEPSSVDKTRAAIRDLHTQITVSIHSVEAISRRIETLRDEELHSQLLELLQGL 516
R+ K + +DKTRA ++DLH++I V+I+SV++IS+RIE +RDEEL+ QLLEL +GL
Sbjct: 187 RHQFAKDQGTQVIDKTRAVVKDLHSRIRVAIYSVDSISKRIEKMRDEELYPQLLELTEGL 366
Query: 517 AKMWKVMAECHQTQKQTLDEA 537
+MWK M ECH Q T+ A
Sbjct: 367 VRMWKAMLECHHAQYITISLA 429
>BI270268 weakly similar to GP|15724286|gb| AT4g35240/F23E12_200 {Arabidopsis
thaliana}, partial (17%)
Length = 634
Score = 104 bits (259), Expect(2) = 5e-24
Identities = 62/164 (37%), Positives = 88/164 (52%)
Frame = +3
Query: 423 STLDRLYAWEKKLYQEVKSGTRVRLAYEKKCLQLRNHDIKGEEPSSVDKTRAAIRDLHTQ 482
STL +L WEKKLY EVK+ ++R +EKKC QLR + K + ++ +A I L T+
Sbjct: 90 STLSKLCMWEKKLYHEVKAEEKLRTLHEKKCKQLRRMNKKDADAQKINSIKAFISILVTK 269
Query: 483 ITVSIHSVEAISRRIETLRDEELHSQLLELLQGLAKMWKVMAECHQTQKQTLDEAKILIA 542
+ VSI V+ IS I L +EEL Q+ + MWK M EC++ Q Q + E K L
Sbjct: 270 MKVSIQVVDKISNSISKLTEEELWPQISRFILMFLGMWKDMQECYKRQYQEIVETKALDV 449
Query: 543 GIDARKQSSMSITDPYRLARSASNLETELRNWRNTFESWITSQK 586
RK + +I +A L+TEL+ W +F WI + K
Sbjct: 450 SSFNRKPCNANID-------AAIKLKTELQKWNLSFLDWIQASK 560
Score = 25.4 bits (54), Expect(2) = 5e-24
Identities = 9/21 (42%), Positives = 14/21 (65%)
Frame = +1
Query: 585 QKSYIHALTGWLLRCMRCEPD 605
QK ++ AL WL+RC+ E +
Sbjct: 556 QKFHVKALNDWLVRCLMYESE 618
>AW696434 similar to GP|15810479|gb| unknown protein {Arabidopsis thaliana},
partial (8%)
Length = 487
Score = 100 bits (249), Expect = 2e-21
Identities = 47/60 (78%), Positives = 55/60 (91%)
Frame = +3
Query: 1 MGCSQSKLDDEEAVKICKDRKRFIKEAVEHRTQFANGHIAYIQSLKRVSAALLDYFEENE 60
MGCSQSKLDDEE+V++CKDRKRFIK+AVE RT+FA GHIAYI+S+KRVSAAL DY E +E
Sbjct: 279 MGCSQSKLDDEESVQLCKDRKRFIKQAVEQRTRFATGHIAYIESMKRVSAALRDYIEGDE 458
>TC83396 similar to GP|22858664|gb|AAN05792.1 unknown {Gossypium hirsutum},
partial (40%)
Length = 921
Score = 91.7 bits (226), Expect = 9e-19
Identities = 57/225 (25%), Positives = 112/225 (49%)
Frame = +3
Query: 438 EVKSGTRVRLAYEKKCLQLRNHDIKGEEPSSVDKTRAAIRDLHTQITVSIHSVEAISRRI 497
+ K+ +++ +EKK +R +++ + +K + + L +Q+ V+ ++++ S I
Sbjct: 69 QTKNAKTIKMEHEKKVELVRKLEMRRADYVKTEKAKKEVEKLESQMMVASQTIDSTSAEI 248
Query: 498 ETLRDEELHSQLLELLQGLAKMWKVMAECHQTQKQTLDEAKILIAGIDARKQSSMSITDP 557
LR+ EL+ QL+EL++GL MW+ M ECHQ QK + + + L S+ ++
Sbjct: 249 VKLREIELYPQLIELVKGLMCMWRSMYECHQVQKHIVQQLEYL-----NTIPSNNPTSEI 413
Query: 558 YRLARSASNLETELRNWRNTFESWITSQKSYIHALTGWLLRCMRCEPDASKLICSPRRSS 617
+R +S LE E++ W +F + + + YI +L+GW LR + + L R++
Sbjct: 414 HR--QSTLQLELEVQQWHQSFCNLFKAHRDYIESLSGW-LRLSLYQFSRNPL----SRTT 572
Query: 618 STHPLFGLCIQWSRRLDALEETAVLDSIDFFAAELGSFYAQQLRE 662
+F LC QW ++ + + + I + + QQ E
Sbjct: 573 EESKIFTLCEQWHLAVEHIPDKVASEGIKSLLTVIHAIVVQQTEE 707
>BF640332 weakly similar to GP|5263327|gb| EST gb|T20649 comes from this
gene. {Arabidopsis thaliana}, partial (12%)
Length = 354
Score = 87.0 bits (214), Expect = 2e-17
Identities = 47/121 (38%), Positives = 66/121 (53%)
Frame = +3
Query: 469 VDKTRAAIRDLHTQITVSIHSVEAISRRIETLRDEELHSQLLELLQGLAKMWKVMAECHQ 528
+D + IR L T+I + I SVE IS RI LRDEEL QL L+ GL +MWK M +CHQ
Sbjct: 6 IDAAESLIRKLLTKINICIKSVETISGRIHKLRDEELQPQLAALINGLIRMWKFMFQCHQ 185
Query: 529 TQKQTLDEAKILIAGIDARKQSSMSITDPYRLARSASNLETELRNWRNTFESWITSQKSY 588
Q Q + E+K ++ Q +++ L EL NW F +W+ ++KSY
Sbjct: 186 KQFQAIMESKTQSLKLNTGLQRDEG-------SKAIIELXRELXNWCTQFNNWVRTKKSY 344
Query: 589 I 589
+
Sbjct: 345 V 347
>TC91170 similar to PIR|D96563|D96563 probable bZIP protein 48652-45869
[imported] - Arabidopsis thaliana, partial (33%)
Length = 1084
Score = 82.8 bits (203), Expect = 4e-16
Identities = 52/171 (30%), Positives = 88/171 (51%), Gaps = 5/171 (2%)
Frame = +2
Query: 343 DLESQFTVVCNAANDVSALLEAKKAQYLSPSNELS-----ASKLLNPVALFRSSPSKIIT 397
DL+ F +A++VS +LEA + Y S + +++++ + RS K I
Sbjct: 170 DLDDHFLKASESAHEVSKMLEATRLHYHSNFADNRGHIDHSARVMRVITWNRSF--KGIP 343
Query: 398 NFSKTRDEVYDDPSEEQCVFSVSHQSTLDRLYAWEKKLYQEVKSGTRVRLAYEKKCLQLR 457
N +D+ D E +H + LD+L AWEKKLY EVK+G ++ Y++K L
Sbjct: 344 NLDDGKDDFDSDEHE-------THATILDKLLAWEKKLYDEVKAGELMKFDYQRKVASLN 502
Query: 458 NHDIKGEEPSSVDKTRAAIRDLHTQITVSIHSVEAISRRIETLRDEELHSQ 508
+G +++K +AA+ LHT+ V + S+++ I LRDE+ S+
Sbjct: 503 RLKKRGNNSEALEKAKAAVSQLHTRYIVDMQSLDSTVSEINRLRDEQFVSK 655
>TC87195 similar to GP|16604629|gb|AAL24107.1 putative bZIP protein
{Arabidopsis thaliana}, partial (38%)
Length = 1412
Score = 72.4 bits (176), Expect = 6e-13
Identities = 54/221 (24%), Positives = 103/221 (46%), Gaps = 1/221 (0%)
Frame = +3
Query: 458 NHDIKGEEPSSVDKTRAAIRDLHTQITVSIHSVEAISRRIETLRDEELHSQLLELLQGLA 517
+ + KG++ + + KT++AI L + I V+ +V S I LRD +L QL+EL G+
Sbjct: 192 SQEYKGDDEAKIFKTKSAINRLQSLIVVTSQAVSTTSTAIIGLRDSDLVPQLVELCHGMM 371
Query: 518 KMWKVMAECHQTQKQTLDEAKILIAGIDARKQSSMSITDPYRLARSASNLETELRNWRNT 577
MW+ M + H+ Q + + + L+ + S ++ +R ++ +LE+ + W ++
Sbjct: 372 YMWRSMHQYHEVQSNIVQQVRGLV-----NRSGGESTSELHR--QATRDLESAVSAWHSS 530
Query: 578 FESWITSQKSYIHALTGWL-LRCMRCEPDASKLICSPRRSSSTHPLFGLCIQWSRRLDAL 636
F I Q+ +I +L GW L M + D + S + F W LD +
Sbjct: 531 FCRLIKFQRDFILSLHGWFKLSLMPVDNDN----VNRMEHSDAYMFFD---DWKLALDRV 689
Query: 637 EETAVLDSIDFFAAELGSFYAQQLREDSAQNATVGSNVNME 677
+T ++I F + ++Q E + T ++ +E
Sbjct: 690 PDTVASEAIKSFINVVHVISSKQAEELKIKKRTENASKELE 812
>AW208157 similar to GP|16604629|gb putative bZIP protein {Arabidopsis
thaliana}, partial (17%)
Length = 419
Score = 69.7 bits (169), Expect = 4e-12
Identities = 35/82 (42%), Positives = 54/82 (65%)
Frame = +2
Query: 423 STLDRLYAWEKKLYQEVKSGTRVRLAYEKKCLQLRNHDIKGEEPSSVDKTRAAIRDLHTQ 482
S+L+RL AWEKKLY+EVKS V++ +EKK L++ + KG++ + + KT++AI L +
Sbjct: 173 SSLERLLAWEKKLYEEVKSREGVKIEHEKKLSSLQSQEYKGDDEAKIFKTKSAINRLQSL 352
Query: 483 ITVSIHSVEAISRRIETLRDEE 504
I V+ +V S I LRD +
Sbjct: 353 IVVTSQAVSTTSTAIIGLRDSD 418
>BG646992 similar to PIR|T06133|T061 hypothetical protein F23E12.200 -
Arabidopsis thaliana, partial (11%)
Length = 761
Score = 67.0 bits (162), Expect = 2e-11
Identities = 77/272 (28%), Positives = 120/272 (43%)
Frame = +1
Query: 175 SQWDSFWNPFTSLDYFGYPNGSSLEQIVMDDENRGLRKVREEEGIPDLEQEEMDDEQEGC 234
S WD F N F + D + YP + + + VREEEGIPDLE E+
Sbjct: 4 STWD-FLNFFENHDKY-YPQTTQYTPTATPSRDSNV--VREEEGIPDLEDEDE------- 150
Query: 235 VVKRNVAEERTKIDVNSSKEEVMVEDVDKHKEEEKEKGTDAETETAQEISDSKINGGECF 294
VVK+ +++ NS+ D H E E+ ++E E + + K+ +
Sbjct: 151 VVKQVHGDQKLVPPPNSNHSN----DHHDHGHLEDEED-ESEVEYEVHVVEKKVVNDDGK 315
Query: 295 QVSKSQSSGHMESSHKEMAIDTEEAKEKTPGFTVYVNRRPENMAEVIKDLESQFTVVCNA 354
SKS+ + PG NR P EV ++++ F ++
Sbjct: 316 PKSKSKPNSAFR-----------------PG-----NRNP---LEVAREIQILFQRASDS 420
Query: 355 ANDVSALLEAKKAQYLSPSNELSASKLLNPVALFRSSPSKIITNFSKTRDEVYDDPSEEQ 414
+ ++ +LE K +Y ++ + SK+L+ VA S S N +++ D D E
Sbjct: 421 GSHIADILEVGKLRY---HHKAATSKMLHLVAPSLSVVSSASRN-AQSGDANSVDLDVEL 588
Query: 415 CVFSVSHQSTLDRLYAWEKKLYQEVKSGTRVR 446
S + STL +LY WEKKLYQEVKS ++R
Sbjct: 589 TTRSRNLSSTLHKLYLWEKKLYQEVKSEEKMR 684
>AW687968 similar to GP|22136982|gb| unknown protein {Arabidopsis thaliana},
partial (9%)
Length = 540
Score = 65.9 bits (159), Expect = 6e-11
Identities = 30/55 (54%), Positives = 38/55 (68%)
Frame = +1
Query: 1 MGCSQSKLDDEEAVKICKDRKRFIKEAVEHRTQFANGHIAYIQSLKRVSAALLDY 55
MGC+QS++D+EE+V CKDRK ++EAV R FA GH Y +LK AAL DY
Sbjct: 187 MGCAQSRIDNEESVSRCKDRKNLMREAVAARNAFAAGHSGYAMALKNTGAALSDY 351
>AJ388762 homologue to PIR|D96563|D965 probable bZIP protein 48652-45869
[imported] - Arabidopsis thaliana, partial (12%)
Length = 617
Score = 49.3 bits (116), Expect = 5e-06
Identities = 33/110 (30%), Positives = 55/110 (50%), Gaps = 5/110 (4%)
Frame = +3
Query: 336 NMAEVIKDLESQFTVVCNAANDVSALLEAKKAQYLSPSNELS-----ASKLLNPVALFRS 390
N+ ++ DL+ F +A++VS +LEA + Y S + +++++ + RS
Sbjct: 147 NLVQIFTDLDDHFLKASESAHEVSKMLEATRLHYHSNFADNRGHIDHSARVMRVITWNRS 326
Query: 391 SPSKIITNFSKTRDEVYDDPSEEQCVFSVSHQSTLDRLYAWEKKLYQEVK 440
K I N +D+ D E +H + LD+L AWEKKLY EV+
Sbjct: 327 F--KGIPNLDDGKDDFDSDEHE-------THATILDKLLAWEKKLYDEVR 449
>BG646203 homologue to PIR|S22697|S226 extensin - Volvox carteri
(fragment), partial (5%)
Length = 684
Score = 45.8 bits (107), Expect = 6e-05
Identities = 27/83 (32%), Positives = 47/83 (56%)
Frame = +3
Query: 16 ICKDRKRFIKEAVEHRTQFANGHIAYIQSLKRVSAALLDYFEENEALEFSFDSFVTPPAK 75
+C+DR +FI E++ A+ H+A++ SL+ + LL +F++ E +FS + T P+
Sbjct: 6 LCRDRFKFIDESLRQSYSLADAHVAHMYSLRTLGPTLLHFFKQFE--DFSGE---TKPSL 170
Query: 76 NKASPAVISISKHSSPTTIEFGP 98
+K+ P S S SS + E P
Sbjct: 171SKSPPPPRSKSSSSSDSDAEHVP 239
>TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein T28I19.100
- Arabidopsis thaliana, partial (14%)
Length = 1460
Score = 42.7 bits (99), Expect = 5e-04
Identities = 49/194 (25%), Positives = 83/194 (42%), Gaps = 18/194 (9%)
Frame = +3
Query: 199 EQIVMDDENRGLRKVREEEGIPDLEQEEMDDEQEGCVVKRN----VAEERTKIDVNSS-- 252
E V+D+E K +EEEG + E E+ +DE++G +V+ + EE K D SS
Sbjct: 684 EDEVVDEE-----KDKEEEGDDETENEDKEDEEKGGLVENHENHEAREEHYKADDASSAV 848
Query: 253 ---KEEVMVE---------DVDKHKEEEKEKGTDAETETAQEISDSKINGGECFQVSKSQ 300
E E + ++ E E E+ +Q +SD K+ GE S
Sbjct: 849 AHDTHETSTETGNLEHSDLSLQNTRKPENETNHSDESYGSQNVSDLKVTEGELTDGVSSN 1028
Query: 301 SSGHMESSHKEMAIDTEEAKEKTPGFTVYVNRRPENMAEVIKDLESQFTVVCNAANDVSA 360
++ E+ + + +T + K P + ++ N+ VI + S + +D S+
Sbjct: 1029ATAGKETGNDSFSNETAKTK---PDSQLDLS---SNLTAVITEASSNSS---GTGDDTSS 1181
Query: 361 LLEAKKAQYLSPSN 374
E KA LS S+
Sbjct: 1182SSEQIKAVILSESD 1223
Score = 38.5 bits (88), Expect = 0.009
Identities = 25/119 (21%), Positives = 52/119 (43%), Gaps = 1/119 (0%)
Frame = +3
Query: 205 DENRGLRKVREEEGIP-DLEQEEMDDEQEGCVVKRNVAEERTKIDVNSSKEEVMVEDVDK 263
D+N G + E+E I +++ + DEQ+ EE K + E+ + E ++
Sbjct: 384 DKNEGHEEEEEDEHIVYNMQNKREHDEQQ------QEGEEGNKHETEEESEDNVHERREE 545
Query: 264 HKEEEKEKGTDAETETAQEISDSKINGGECFQVSKSQSSGHMESSHKEMAIDTEEAKEK 322
EEE + G + + E + + + GG+ ++ ++ +D E+ KE+
Sbjct: 546 QDEEENKHGAEVQEENESKSEEVEDEGGDVEIDENDHEKSEADNDREDEVVDEEKDKEE 722
>TC88896 weakly similar to GP|6728968|gb|AAF26966.1| unknown protein
{Arabidopsis thaliana}, partial (27%)
Length = 1097
Score = 40.8 bits (94), Expect = 0.002
Identities = 61/285 (21%), Positives = 114/285 (39%), Gaps = 3/285 (1%)
Frame = +2
Query: 225 EEMDDEQEGCVVKRNVAEERTKIDVNSSKEEVMVEDVDKHKEEEKEKGTDAETETAQ-EI 283
E+++ E E + + A R+ +D K E + V++ + E+ ET T Q E
Sbjct: 95 EQLNVESEAAKMAESYA--RSVLDECRKKVEELEMKVEEANQLERSASLSLETATKQLEG 268
Query: 284 SDSKINGGECFQVSKSQSSGHMESSHKEMAIDTEEAKEKTPGFTVYVNRRPEN--MAEVI 341
+ ++ E S + G +E + D E+A+ + + EN M++ I
Sbjct: 269 KNELLHDAESEISSLKEKLGMLEMTVGRQRGDLEDAER------CLLAAKEENIEMSKKI 430
Query: 342 KDLESQFTVVCNAANDVSALLEAKKAQYLSPSNELSASKLLNPVALFRSSPSKIITNFSK 401
+ LES+ V +KAQ L+ + +LSAS V +K+I
Sbjct: 431 ESLESEIETVSK-----------EKAQALN-NEKLSASS----VQTLLEEKNKLINELEI 562
Query: 402 TRDEVYDDPSEEQCVFSVSHQSTLDRLYAWEKKLYQEVKSGTRVRLAYEKKCLQLRNHDI 461
RDE + S H+ + + EK L + E + + + D+
Sbjct: 563 CRDEEEKTKLAMDSLASALHEVSAEARDTKEKLLANQA----------EHESYETQIEDL 712
Query: 462 KGEEPSSVDKTRAAIRDLHTQITVSIHSVEAISRRIETLRDEELH 506
K + +S +K + + D H +I +EA + E++ ++ H
Sbjct: 713 KSDLEASKEKYESMLNDAHHEIEDLKSDLEASKEKYESMLNDAHH 847
>TC83625 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (9%)
Length = 908
Score = 39.7 bits (91), Expect = 0.004
Identities = 31/122 (25%), Positives = 55/122 (44%)
Frame = +2
Query: 201 IVMDDENRGLRKVREEEGIPDLEQEEMDDEQEGCVVKRNVAEERTKIDVNSSKEEVMVED 260
I +DD++ G K + E DLE + ++ + E K +++T +D K +D
Sbjct: 479 IKIDDKSAGDVKEDKVEIEKDLEIKSVEKDDEKKEKKDKEKKDKTDVDEGKDK-----KD 643
Query: 261 VDKHKEEEKEKGTDAETETAQEISDSKINGGECFQVSKSQSSGHMESSHKEMAIDTEEAK 320
+K K+E+KE+ E E E D + E + K + + K+ D E+ K
Sbjct: 644 KEKKKKEKKEENVKGEEEDGDEKKDKEKKKKE--KKEKGKEDKDKDGEEKKSKKDKEKKK 817
Query: 321 EK 322
+K
Sbjct: 818 DK 823
>TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (13%)
Length = 663
Score = 39.7 bits (91), Expect = 0.004
Identities = 31/122 (25%), Positives = 55/122 (44%)
Frame = +3
Query: 201 IVMDDENRGLRKVREEEGIPDLEQEEMDDEQEGCVVKRNVAEERTKIDVNSSKEEVMVED 260
I +DD++ G K + E DLE + ++ + E K +++T +D K +D
Sbjct: 87 IKIDDKSAGDVKEDKVEIEKDLEIKSVEKDDEKKEKKDKEKKDKTDVDEGKDK-----KD 251
Query: 261 VDKHKEEEKEKGTDAETETAQEISDSKINGGECFQVSKSQSSGHMESSHKEMAIDTEEAK 320
+K K+E+KE+ E E E D + E + K + + K+ D E+ K
Sbjct: 252 KEKKKKEKKEENVKGEEEDGDEKKDKEKKKKE--KKEKGKEDKDKDGEEKKSKKDKEKKK 425
Query: 321 EK 322
+K
Sbjct: 426 DK 431
Score = 29.6 bits (65), Expect = 4.4
Identities = 24/89 (26%), Positives = 40/89 (43%), Gaps = 7/89 (7%)
Frame = +3
Query: 204 DDENRGLRKVREEEGIPDLEQEEMDDEQEGCVVKRNVAEERTKIDVNSSKEE--VMVEDV 261
D + G K +++ ++ E DDE E K+ +++ K KEE V V D+
Sbjct: 369 DKDKDGEEKKSKKDKEKKKDKNEDDDEGEDGSKKKKNKDKKEKKKEEDEKEEGKVSVRDI 548
Query: 262 D-----KHKEEEKEKGTDAETETAQEISD 285
D K +E+K+K D E + + D
Sbjct: 549 DIEETAKEGKEKKKKKEDKEEKKKKSGKD 635
>TC86635 weakly similar to GP|9758600|dbj|BAB09233.1
gene_id:MDF20.10~pir||T08929~similar to unknown protein
{Arabidopsis thaliana}, partial (32%)
Length = 1963
Score = 37.0 bits (84), Expect = 0.027
Identities = 22/74 (29%), Positives = 41/74 (54%), Gaps = 1/74 (1%)
Frame = +2
Query: 204 DDENRGLRKVREEEGIPDLEQEEMDDEQEGCVVKRNVAEERTKIDVNSSKEEVMVED-VD 262
+DE + + +EE + +EE ++++EG +E+ K + + +EE ED +D
Sbjct: 17 NDEEKTEVEEKEEGDDNEKSEEETEEKEEG--------DEKEKTEAETEEEEEADEDTID 172
Query: 263 KHKEEEKEKGTDAE 276
K KEE+K +G+ E
Sbjct: 173 KSKEEDKAEGSKGE 214
Score = 36.2 bits (82), Expect = 0.047
Identities = 31/99 (31%), Positives = 48/99 (48%)
Frame = +2
Query: 224 QEEMDDEQEGCVVKRNVAEERTKIDVNSSKEEVMVEDVDKHKEEEKEKGTDAETETAQEI 283
QEE +DE+ + EE+ + D N EE E +K + +EKEK T+AETE +E
Sbjct: 5 QEEGNDEE------KTEVEEKEEGDDNEKSEE---ETEEKEEGDEKEK-TEAETEEEEEA 154
Query: 284 SDSKINGGECFQVSKSQSSGHMESSHKEMAIDTEEAKEK 322
+ I+ + + K++ S + S K E K K
Sbjct: 155 DEDTIDKSK--EEDKAEGSKGEKGSKKRARGKVNEEKVK 265
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 37.0 bits (84), Expect = 0.027
Identities = 23/82 (28%), Positives = 43/82 (52%)
Frame = +3
Query: 204 DDENRGLRKVREEEGIPDLEQEEMDDEQEGCVVKRNVAEERTKIDVNSSKEEVMVEDVDK 263
++E+ G +V EEE D E+E+ DE+E EE + D + +EE E+ ++
Sbjct: 57 EEEDEG--EVEEEEDEHDEEEEDEHDEEE------EEEEEEEEEDEHDDEEEEEEEEEEE 212
Query: 264 HKEEEKEKGTDAETETAQEISD 285
+E++ E+G + E + + D
Sbjct: 213 EEEDDDEEGEEDEIDRISTLPD 278
>TC80941 weakly similar to PIR|T39903|T39903 serine-rich protein - fission
yeast (Schizosaccharomyces pombe), partial (15%)
Length = 557
Score = 37.0 bits (84), Expect = 0.027
Identities = 25/85 (29%), Positives = 45/85 (52%), Gaps = 1/85 (1%)
Frame = +2
Query: 195 GSSLEQIVMDDENRGLRKVREEEGIPDLEQEEMDDEQEGCVVKRNVAEERTKIDVNSSKE 254
GS E+ +DE + + +EE + +EE ++++EG +E+ K + + +E
Sbjct: 149 GSIEEKEEGNDEEKTEVEEKEEGDDNEKSEEETEEKEEG--------DEKEKTEAETEEE 304
Query: 255 EVMVED-VDKHKEEEKEKGTDAETE 278
E ED +DK KEE+K +G+ E
Sbjct: 305 EEADEDTIDKSKEEDKAEGSKGGKE 379
Score = 36.6 bits (83), Expect = 0.036
Identities = 28/85 (32%), Positives = 45/85 (52%)
Frame = +2
Query: 205 DENRGLRKVREEEGIPDLEQEEMDDEQEGCVVKRNVAEERTKIDVNSSKEEVMVEDVDKH 264
+E + +E EG + E+EE +DE+ + EE+ + D N EE E +K
Sbjct: 110 EEKEKIEAEKEAEGSIE-EKEEGNDEE------KTEVEEKEEGDDNEKSEE---ETEEKE 259
Query: 265 KEEEKEKGTDAETETAQEISDSKIN 289
+ +EKEK T+AETE +E + I+
Sbjct: 260 EGDEKEK-TEAETEEEEEADEDTID 331
>TC86517 homologue to PIR|T09640|T09640 protein phosphatase 2C - alfalfa,
complete
Length = 1616
Score = 33.9 bits (76), Expect = 0.23
Identities = 25/75 (33%), Positives = 37/75 (49%)
Frame = -3
Query: 209 GLRKVREEEGIPDLEQEEMDDEQEGCVVKRNVAEERTKIDVNSSKEEVMVEDVDKHKEEE 268
G R+ + EEG E EE DDE VV+ + ER + V V D+ + ++EE
Sbjct: 327 GNRRQKGEEGEEQGEGEEEDDEDVVVVVEDGLR*ER---------DNVNVSDLLQKRDEE 175
Query: 269 KEKGTDAETETAQEI 283
E + ET T Q++
Sbjct: 174 GENTGEFETATEQDM 130
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.313 0.128 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,686,918
Number of Sequences: 36976
Number of extensions: 282585
Number of successful extensions: 1453
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 1395
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1429
length of query: 738
length of database: 9,014,727
effective HSP length: 103
effective length of query: 635
effective length of database: 5,206,199
effective search space: 3305936365
effective search space used: 3305936365
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC125477.15


