

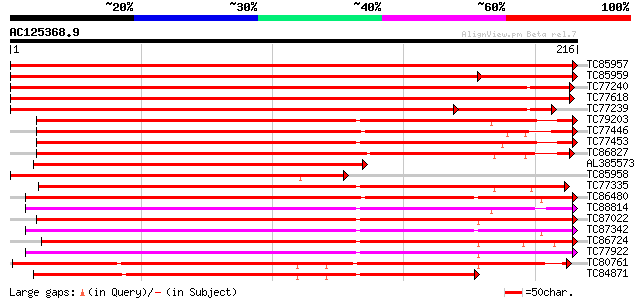
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125368.9 + phase: 0
(216 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85957 homologue to PIR|S57471|S57471 GTP-binding protein GTP6 ... 413 e-116
TC85959 homologue to PIR|S57471|S57471 GTP-binding protein GTP6 ... 355 e-115
TC77240 homologue to GP|1370196|emb|CAA98175.1 RAB8D {Lotus japo... 405 e-114
TC77618 homologue to PIR|S57478|S57478 GTP-binding protein GTP13... 402 e-113
TC77239 homologue to GP|1370196|emb|CAA98175.1 RAB8D {Lotus japo... 339 e-105
TC79203 homologue to PIR|S41430|S41430 GTP-binding protein ras-... 249 6e-67
TC77446 homologue to GP|303750|dbj|BAA02116.1| GTP-binding prote... 245 1e-65
TC77453 homologue to GP|303732|dbj|BAA02117.1| GTP-binding prote... 242 7e-65
TC86827 homologue to GP|303748|dbj|BAA02115.1| GTP-binding prote... 230 3e-61
AL385573 homologue to GP|11558647|em secretion related GTPase (... 226 5e-60
TC85958 PIR|S57471|S57471 GTP-binding protein GTP6 - garden pea,... 207 2e-54
TC77335 homologue to GP|1370176|emb|CAA98165.1 RAB2A {Lotus japo... 181 1e-46
TC86480 homologue to SP|Q40193|R11C_LOTJA Ras-related protein Ra... 181 1e-46
TC88814 homologue to PIR|T06447|T06447 GTP-binding protein - gar... 177 2e-45
TC87022 homologue to SP|O04486|RB1C_ARATH Ras-related protein Ra... 176 6e-45
TC87342 PIR|S41431|S41431 GTP-binding protein ras-like - fava b... 172 9e-44
TC86724 homologue to PIR|T06445|T06445 GTP-binding protein - gar... 169 6e-43
TC77922 homologue to PIR|T06448|T06448 GTP-binding protein - gar... 167 2e-42
TC80761 homologue to GP|1370174|emb|CAA98164.1 RAB1Y {Lotus japo... 167 4e-42
TC84871 similar to GP|1370172|emb|CAA98163.1 RAB1X {Lotus japoni... 166 5e-42
>TC85957 homologue to PIR|S57471|S57471 GTP-binding protein GTP6 - garden
pea, complete
Length = 1148
Score = 413 bits (1061), Expect = e-116
Identities = 204/216 (94%), Positives = 215/216 (99%)
Frame = +3
Query: 1 MAAPPARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD 60
MAAPPARAR+DYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD
Sbjct: 207 MAAPPARARSDYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD 386
Query: 61 GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDN 120
GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNI+NWIRNIEQHASDN
Sbjct: 387 GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIKNWIRNIEQHASDN 566
Query: 121 VNKILVGNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQR 180
VNKILVGNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTN+NV+EVFFSIARDIKQR
Sbjct: 567 VNKILVGNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNMNVDEVFFSIARDIKQR 746
Query: 181 LADTDSKSEPQTIKINQPDQGAGAAQAAQKSACCGS 216
LA++DSK+EPQT+KINQPDQGAG+AQA+Q S+CCGS
Sbjct: 747 LAESDSKTEPQTLKINQPDQGAGSAQASQTSSCCGS 854
>TC85959 homologue to PIR|S57471|S57471 GTP-binding protein GTP6 - garden
pea, complete
Length = 886
Score = 355 bits (912), Expect(2) = e-115
Identities = 179/180 (99%), Positives = 180/180 (99%)
Frame = +3
Query: 1 MAAPPARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD 60
MAAPPARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD
Sbjct: 72 MAAPPARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD 251
Query: 61 GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDN 120
GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDN
Sbjct: 252 GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDN 431
Query: 121 VNKILVGNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQR 180
VNKILVGNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQ+
Sbjct: 432 VNKILVGNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQK 611
Score = 78.2 bits (191), Expect(2) = e-115
Identities = 37/38 (97%), Positives = 38/38 (99%)
Frame = +1
Query: 179 QRLADTDSKSEPQTIKINQPDQGAGAAQAAQKSACCGS 216
+RLADTDSKSEPQTIKINQPDQGAGAAQAAQKSACCGS
Sbjct: 607 KRLADTDSKSEPQTIKINQPDQGAGAAQAAQKSACCGS 720
>TC77240 homologue to GP|1370196|emb|CAA98175.1 RAB8D {Lotus japonicus},
complete
Length = 1148
Score = 405 bits (1040), Expect = e-114
Identities = 205/215 (95%), Positives = 209/215 (96%)
Frame = +1
Query: 1 MAAPPARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD 60
MAAPPARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD
Sbjct: 223 MAAPPARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD 402
Query: 61 GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDN 120
GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDE+SFNNIRNWIRNIEQHASDN
Sbjct: 403 GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDN 582
Query: 121 VNKILVGNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQR 180
VNKILVGNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQR
Sbjct: 583 VNKILVGNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQR 762
Query: 181 LADTDSKSEPQTIKINQPDQGAGAAQAAQKSACCG 215
LADTD+K+EP TIKINQ D G+ QAAQKSACCG
Sbjct: 763 LADTDNKAEPTTIKINQ-DSATGSGQAAQKSACCG 864
>TC77618 homologue to PIR|S57478|S57478 GTP-binding protein GTP13 - garden
pea, complete
Length = 1136
Score = 402 bits (1034), Expect = e-113
Identities = 203/215 (94%), Positives = 207/215 (95%)
Frame = +2
Query: 1 MAAPPARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD 60
MAA PARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD
Sbjct: 218 MAAAPARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD 397
Query: 61 GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDN 120
GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDE+SFNNIRNWIRNIEQHASDN
Sbjct: 398 GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDN 577
Query: 121 VNKILVGNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQR 180
VNKILVGNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTN+NVEEVFFSIARDIKQR
Sbjct: 578 VNKILVGNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNMNVEEVFFSIARDIKQR 757
Query: 181 LADTDSKSEPQTIKINQPDQGAGAAQAAQKSACCG 215
LADT S +EPQ+IKINQ DQ A QAAQKSACCG
Sbjct: 758 LADTVSSTEPQSIKINQQDQAANGGQAAQKSACCG 862
>TC77239 homologue to GP|1370196|emb|CAA98175.1 RAB8D {Lotus japonicus},
partial (96%)
Length = 816
Score = 339 bits (870), Expect(2) = e-105
Identities = 170/171 (99%), Positives = 171/171 (99%)
Frame = +1
Query: 1 MAAPPARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD 60
MAAPPARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD
Sbjct: 196 MAAPPARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD 375
Query: 61 GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDN 120
GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDE+SFNNIRNWIRNIEQHASDN
Sbjct: 376 GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDN 555
Query: 121 VNKILVGNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFF 171
VNKILVGNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFF
Sbjct: 556 VNKILVGNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFF 708
Score = 58.5 bits (140), Expect(2) = e-105
Identities = 30/39 (76%), Positives = 33/39 (83%)
Frame = +3
Query: 170 FFSIARDIKQRLADTDSKSEPQTIKINQPDQGAGAAQAA 208
FFSIARDIKQRLADTD+K+EP TIKINQ D G+ QAA
Sbjct: 702 FFSIARDIKQRLADTDNKAEPTTIKINQ-DSATGSGQAA 815
>TC79203 homologue to PIR|S41430|S41430 GTP-binding protein ras-like (clone
vfa-ypt1) - fava bean, complete
Length = 920
Score = 249 bits (636), Expect = 6e-67
Identities = 126/207 (60%), Positives = 156/207 (74%), Gaps = 1/207 (0%)
Frame = +1
Query: 11 DYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWD 70
+YDYL KLLLIGDSGVGKSCLLLRF+D S+ S+I+TIG+DFKIRT+E DGK IKLQIWD
Sbjct: 217 EYDYLFKLLLIGDSGVGKSCLLLRFADDSYIDSYISTIGVDFKIRTVEQDGKTIKLQIWD 396
Query: 71 TAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKA 130
TAGQERFRTIT++YYRGA GI++VYDVTDE SFNN++ W+ I+++ASDNVNK+LVGNK
Sbjct: 397 TAGQERFRTITSSYYRGAHGIIIVYDVTDEESFNNVKQWLSEIDRYASDNVNKLLVGNKC 576
Query: 131 DMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLA-DTDSKSE 189
D+ S+RAV +A ADE GI F ETSAK + NVE+ F ++A IK R+A + +
Sbjct: 577 DL-TSERAVSYDTAKAFADEIGIPFMETSAKDSTNVEQAFMAMASSIKDRMASQPTNNAR 753
Query: 190 PQTIKINQPDQGAGAAQAAQKSACCGS 216
P T++I G QKS CC S
Sbjct: 754 PPTVQIRGQPVG-------QKSGCCSS 813
>TC77446 homologue to GP|303750|dbj|BAA02116.1| GTP-binding protein {Pisum
sativum}, complete
Length = 1080
Score = 245 bits (625), Expect = 1e-65
Identities = 123/208 (59%), Positives = 158/208 (75%), Gaps = 2/208 (0%)
Frame = +3
Query: 11 DYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWD 70
+YDYL KLLLIGDSGVGKSCLLLRF+D S+ S+I+TIG+DFKIRT+E DGK IKLQIWD
Sbjct: 126 EYDYLFKLLLIGDSGVGKSCLLLRFADDSYLDSYISTIGVDFKIRTVEQDGKTIKLQIWD 305
Query: 71 TAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKA 130
TAGQERFRTIT++YYRGA GI++VYDVTD+ SFNN++ W+ I+++AS+NVNK+LVGNK+
Sbjct: 306 TAGQERFRTITSSYYRGAHGIIVVYDVTDQESFNNVKQWLNEIDRYASENVNKLLVGNKS 485
Query: 131 DMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDSKS-E 189
D+ + K+ V + +A ADE GI F ETSAK NVE+ F ++A +IK R+A + S
Sbjct: 486 DLSD-KKVVSSETAKAFADEIGIPFMETSAKNASNVEQAFMAMAAEIKNRMASQPANSAR 662
Query: 190 PQTIKI-NQPDQGAGAAQAAQKSACCGS 216
P T++I QP QK+ CC S
Sbjct: 663 PATVQIRGQP--------VNQKAGCCSS 722
>TC77453 homologue to GP|303732|dbj|BAA02117.1| GTP-binding protein {Pisum
sativum}, complete
Length = 944
Score = 242 bits (618), Expect = 7e-65
Identities = 124/208 (59%), Positives = 153/208 (72%), Gaps = 2/208 (0%)
Frame = +1
Query: 11 DYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWD 70
+YDYL KLLLIGDSGVGKSCLLLRF+D S+ S+I+TIG+DFKIRT+E DGK IKLQIWD
Sbjct: 133 EYDYLFKLLLIGDSGVGKSCLLLRFADDSYIDSYISTIGVDFKIRTVEQDGKTIKLQIWD 312
Query: 71 TAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKA 130
TAGQERFRTIT++YYRGA GI++VYDVTDE SFNN++ W+ I+++ASDNVNK+LVGNK+
Sbjct: 313 TAGQERFRTITSSYYRGAHGIIIVYDVTDEESFNNVKQWLSEIDRYASDNVNKLLVGNKS 492
Query: 131 DMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDS--KS 188
D+ + RAV + AD+ GI F ETSAK NVE F ++A IK R+A S +
Sbjct: 493 DL-TANRAVSYDTAKEFADQIGIPFMETSAKDATNVEGAFMAMAAAIKDRMASQPSANNA 669
Query: 189 EPQTIKINQPDQGAGAAQAAQKSACCGS 216
P T++I G QKS CC S
Sbjct: 670 RPPTVQIRGQPVG-------QKSGCCSS 732
>TC86827 homologue to GP|303748|dbj|BAA02115.1| GTP-binding protein {Pisum
sativum}, complete
Length = 917
Score = 230 bits (587), Expect = 3e-61
Identities = 115/208 (55%), Positives = 157/208 (75%), Gaps = 3/208 (1%)
Frame = +2
Query: 11 DYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWD 70
+YDYL KLL+IGDS VGKSCLLLRF+D S+ ++I+TIG+DFKIRT+EL+GK KLQIWD
Sbjct: 134 EYDYLFKLLIIGDSSVGKSCLLLRFADDSYDDTYISTIGVDFKIRTVELEGKTAKLQIWD 313
Query: 71 TAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKA 130
TAGQERFRTIT++YYRGA GI++VYDVTD SFNN++ W+ I+++A+ +V+K+LVGNK
Sbjct: 314 TAGQERFRTITSSYYRGAHGIIIVYDVTDIESFNNVKQWLHEIDRYANHSVSKLLVGNKC 493
Query: 131 DMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLAD--TDSKS 188
D+ ++K V T +A ADE GI F ETSAK ++NVE+ F ++A +IK ++ T SKS
Sbjct: 494 DLTDNK-LVHTHTAKAFADELGIPFLETSAKDSINVEQAFLTMAAEIKNKMGSQPTGSKS 670
Query: 189 EPQTIKI-NQPDQGAGAAQAAQKSACCG 215
+++++ QP Q Q + CCG
Sbjct: 671 AAESVQMKGQPIQ--------QNTNCCG 730
>AL385573 homologue to GP|11558647|em secretion related GTPase (SrgA)
{Aspergillus niger}, partial (61%)
Length = 463
Score = 226 bits (576), Expect = 5e-60
Identities = 113/127 (88%), Positives = 115/127 (89%)
Frame = +2
Query: 10 ADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIW 69
A YDYLIKLLLIGDSGVGKSCLLLRFSD SFT SFITTIGIDFKIRTIELDGKRIKLQIW
Sbjct: 74 AAYDYLIKLLLIGDSGVGKSCLLLRFSDDSFTPSFITTIGIDFKIRTIELDGKRIKLQIW 253
Query: 70 DTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNK 129
DTAGQERFRTITTAYYRGAMGILLVYDVTDE SF NIRNW NIEQHAS+ VNKIL+GNK
Sbjct: 254 DTAGQERFRTITTAYYRGAMGILLVYDVTDERSFKNIRNWFSNIEQHASEGVNKILIGNK 433
Query: 130 ADMDESK 136
D E K
Sbjct: 434 CDWVEKK 454
>TC85958 PIR|S57471|S57471 GTP-binding protein GTP6 - garden pea, partial
(59%)
Length = 646
Score = 207 bits (527), Expect = 2e-54
Identities = 109/130 (83%), Positives = 113/130 (86%), Gaps = 1/130 (0%)
Frame = +2
Query: 1 MAAPPARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD 60
MAAPPARAR+DYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD
Sbjct: 239 MAAPPARARSDYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD 418
Query: 61 GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNW-IRNIEQHASD 119
GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDE F + + SD
Sbjct: 419 GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDEVVF*QHQELDSATLSSMLSD 598
Query: 120 NVNKILVGNK 129
NVNKIL G +
Sbjct: 599 NVNKILGGEQ 628
Score = 35.8 bits (81), Expect = 0.013
Identities = 22/38 (57%), Positives = 24/38 (62%), Gaps = 4/38 (10%)
Frame = +3
Query: 101 SSFNNIRNWIRNIEQHAS----DNVNKILVGNKADMDE 134
SSFNNI+NWIR QH + K VGNKADMDE
Sbjct: 540 SSFNNIKNWIR---QH*AACFLTMSTKY*VGNKADMDE 644
>TC77335 homologue to GP|1370176|emb|CAA98165.1 RAB2A {Lotus japonicus},
complete
Length = 1121
Score = 181 bits (460), Expect = 1e-46
Identities = 98/209 (46%), Positives = 129/209 (60%), Gaps = 7/209 (3%)
Frame = +3
Query: 12 YDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDT 71
Y YL K ++IGD+GVGKSCLLL+F+D F TIG++F R I +D K IKLQIWDT
Sbjct: 237 YAYLFKYIIIGDTGVGKSCLLLQFTDKRFQPVHDLTIGVEFGARMITIDNKPIKLQIWDT 416
Query: 72 AGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKAD 131
AGQE FR+IT +YYRGA G LLVYD+T +FN++ +W+ + QHA+ N+ +L+GNK D
Sbjct: 417 AGQESFRSITRSYYRGAAGALLVYDITRRETFNHLASWLEDARQHANANMTIMLIGNKCD 596
Query: 132 MDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLAD--TDSKSE 189
+ +RAV T +G+ A E G+ F E SAKT NVEE F A I +++ D D +E
Sbjct: 597 L-AHRRAVSTEEGEQFAKENGLIFMEASAKTAQNVEEAFIKTAGTIYKKIQDGVFDVSNE 773
Query: 190 PQTIKINQ-----PDQGAGAAQAAQKSAC 213
IK+ P G AA C
Sbjct: 774 SYGIKVGYGGIPGPSGGRDGPSAAGGGCC 860
>TC86480 homologue to SP|Q40193|R11C_LOTJA Ras-related protein Rab11C.
{Lotus japonicus}, complete
Length = 1173
Score = 181 bits (460), Expect = 1e-46
Identities = 92/214 (42%), Positives = 134/214 (61%), Gaps = 4/214 (1%)
Frame = +3
Query: 7 RARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKL 66
R +YDYL K++LIGDSGVGKS +L RF+ F +TIG++F RT++++GK +K
Sbjct: 204 RVDHEYDYLFKIVLIGDSGVGKSNILSRFTRNEFCLESKSTIGVEFATRTLQVEGKTVKA 383
Query: 67 QIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILV 126
QIWDTAGQER+R IT+AYYRGA+G LLVYD+T +F+N+ W+R + HA N+ ++
Sbjct: 384 QIWDTAGQERYRAITSAYYRGAVGALLVYDITKRQTFDNVNRWLRELRDHADSNIVIMMA 563
Query: 127 GNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDS 186
GNK+D++ RAV GQALA++ G+ F ETSA N+E+ F +I +I + +
Sbjct: 564 GNKSDLNH-LRAVSEDDGQALAEKEGLSFLETSALEATNIEKAFQTILTEI-YHIVSKKA 737
Query: 187 KSEPQTIKINQPDQG----AGAAQAAQKSACCGS 216
+ + + P QG + A K CC +
Sbjct: 738 LAAQEAAGTSLPGQGTTINVADSSANTKRGCCST 839
>TC88814 homologue to PIR|T06447|T06447 GTP-binding protein - garden pea,
complete
Length = 1183
Score = 177 bits (450), Expect = 2e-45
Identities = 97/218 (44%), Positives = 130/218 (59%), Gaps = 8/218 (3%)
Frame = +3
Query: 7 RARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKL 66
RA DYDYL K++LIGDSGVGKS LL RF+ F+ +TIG++F R+I +D K +K
Sbjct: 297 RADDDYDYLFKVVLIGDSGVGKSNLLSRFTKNEFSLESKSTIGVEFATRSIRVDDKVVKA 476
Query: 67 QIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILV 126
QIWDTAGQER+R IT+AYYRGA+G LLVYDVT +F N+ W++ + H N+ +LV
Sbjct: 477 QIWDTAGQERYRAITSAYYRGAVGALLVYDVTRHVTFENVERWLKELRDHTDANIVVMLV 656
Query: 127 GNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLA---- 182
GNKAD+ RAV T A A+ F ETSA ++NVE F + I + ++
Sbjct: 657 GNKADL-RHLRAVSTEDSTAFAERENTFFMETSALESMNVENAFTEVLTQIYRVVSKKAL 833
Query: 183 ----DTDSKSEPQTIKINQPDQGAGAAQAAQKSACCGS 216
D + + QTI + D A +KS CC +
Sbjct: 834 EIGDDPTALPKGQTIDVGSRDD----VSAVKKSGCCSA 935
>TC87022 homologue to SP|O04486|RB1C_ARATH Ras-related protein Rab11C.
[Mouse-ear cress] {Arabidopsis thaliana}, partial (81%)
Length = 1014
Score = 176 bits (446), Expect = 6e-45
Identities = 91/211 (43%), Positives = 132/211 (62%), Gaps = 5/211 (2%)
Frame = +2
Query: 11 DYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWD 70
+YDYL K++LIGDSGVGKS LL RF+ F +TIG++F RT++++G+ +K QIWD
Sbjct: 74 EYDYLFKVVLIGDSGVGKSNLLSRFTRNEFCLESKSTIGVEFATRTLQVEGRTVKAQIWD 253
Query: 71 TAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKA 130
TAGQER+R IT+AYYRGA+G LLVYDVT ++F+N+ W++ + HA N+ +L+GNK
Sbjct: 254 TAGQERYRAITSAYYRGALGALLVYDVTKPTTFDNVTRWLKELRDHADANIVIMLIGNKT 433
Query: 131 DMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDI-----KQRLADTD 185
D+ + RAV T Q+ A++ G+ F ETSA NVE+ F + +I K+ L+ +
Sbjct: 434 DL-KHLRAVATEDAQSYAEKEGLSFIETSALEATNVEKAFQTTLGEIYRIISKKSLSTAN 610
Query: 186 SKSEPQTIKINQPDQGAGAAQAAQKSACCGS 216
+ IK + G+ CC S
Sbjct: 611 EPAAAANIKEGKTIAIGGSETTNTNKPCCTS 703
>TC87342 PIR|S41431|S41431 GTP-binding protein ras-like - fava bean,
complete
Length = 995
Score = 172 bits (436), Expect = 9e-44
Identities = 94/215 (43%), Positives = 127/215 (58%), Gaps = 5/215 (2%)
Frame = +3
Query: 7 RARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKL 66
RA DYDYL K++LIGDSGVGKS LL RF+ F +TIG++F RT+ +D K IK
Sbjct: 87 RADDDYDYLFKVVLIGDSGVGKSNLLSRFTKNEFNLESKSTIGVEFATRTLNVDSKVIKA 266
Query: 67 QIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILV 126
QIWDTAGQER+R IT+AYYRGA+G LLVYDVT ++F N+ W++ + H N+ +LV
Sbjct: 267 QIWDTAGQERYRAITSAYYRGAVGALLVYDVTRHATFENVDRWLKELRNHTDSNIVVMLV 446
Query: 127 GNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDS 186
GNKAD+ AV T G++ A++ + F ETSA NVE F + I R+ +
Sbjct: 447 GNKADL-RHLVAVSTEDGKSYAEKESLYFMETSALEATNVENAFAEVLTQI-YRIVSKKA 620
Query: 187 KSEPQTIKINQPDQG-----AGAAQAAQKSACCGS 216
+ + P +G A ++ CC S
Sbjct: 621 VEGAENGNASVPAKGEKIDLKNDVSALKRVGCCSS 725
>TC86724 homologue to PIR|T06445|T06445 GTP-binding protein - garden pea,
complete
Length = 1037
Score = 169 bits (429), Expect = 6e-43
Identities = 92/212 (43%), Positives = 134/212 (62%), Gaps = 8/212 (3%)
Frame = +3
Query: 13 DYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTA 72
DY+ K++LIGDS VGKS +L RFS F+ +TIG++F+ RT+ +D K +K QIWDTA
Sbjct: 216 DYVFKVVLIGDSAVGKSQILARFSRNEFSLDSKSTIGVEFQTRTLVIDHKTVKAQIWDTA 395
Query: 73 GQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKADM 132
GQER+R +T+AYYRGA+G +LVYD+T +F++I W+ + HA N+ ILVGNK+D+
Sbjct: 396 GQERYRAVTSAYYRGAVGAMLVYDITKRQTFDHIPRWLEELRNHADKNIVIILVGNKSDL 575
Query: 133 DESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDI-----KQRLADTDSK 187
E++R VPT + A++ G+ F ETSA NVE F ++ +I K+ LA +S+
Sbjct: 576 -ENQRDVPTEDAKEFAEKEGLFFLETSALQATNVEASFMTVLTEIYNIVNKKNLAADESQ 752
Query: 188 SEPQTIK-INQPDQGAGAAQ--AAQKSACCGS 216
+ + Q G AQ A+ + CC S
Sbjct: 753 GNGNSASLLGQKIIIPGPAQEIPAKSNMCCQS 848
>TC77922 homologue to PIR|T06448|T06448 GTP-binding protein - garden pea,
complete
Length = 1182
Score = 167 bits (424), Expect = 2e-42
Identities = 89/215 (41%), Positives = 125/215 (57%), Gaps = 5/215 (2%)
Frame = +2
Query: 7 RARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKL 66
RA +YDYL KL+LIGDSGVGKS LL RF+ F +TIG++F +++ +D K IK
Sbjct: 176 RADDEYDYLFKLVLIGDSGVGKSNLLSRFTRNEFNLESKSTIGVEFATKSLTVDAKVIKA 355
Query: 67 QIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILV 126
QIWDTAGQER+R IT+AYYRGA+G LLVYDVT ++F N W++ + H N+ +L+
Sbjct: 356 QIWDTAGQERYRAITSAYYRGAVGALLVYDVTRRATFENAARWLKELRDHTDPNIVVMLI 535
Query: 127 GNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDI-----KQRL 181
GNK+D+ AVPT G++ A+ + F ETSA NVE F + I K+ +
Sbjct: 536 GNKSDL-RHLVAVPTEDGKSFAERESLYFMETSALEATNVENAFTEVLSQIYRIVSKKAV 712
Query: 182 ADTDSKSEPQTIKINQPDQGAGAAQAAQKSACCGS 216
+ S + Q + ++ CC +
Sbjct: 713 EAGEGGSSSSVPSVGQTINVKEDSSVLKRFGCCSN 817
>TC80761 homologue to GP|1370174|emb|CAA98164.1 RAB1Y {Lotus japonicus},
partial (94%)
Length = 994
Score = 167 bits (422), Expect = 4e-42
Identities = 94/217 (43%), Positives = 133/217 (60%), Gaps = 4/217 (1%)
Frame = +1
Query: 2 AAPPARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDG 61
A + + ++DY+ KLL+IGDSGVGKS LLLRF+ F TIG+DFK++ + ++G
Sbjct: 109 AMDSSSGQQEFDYMFKLLMIGDSGVGKSSLLLRFTSDDFD-DLSPTIGVDFKVKYVTIEG 285
Query: 62 KRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRN-WIRNIEQHASD- 119
K++KL IWDTAGQERFRT+T++YYRGA GI++VYDVT +F N+ W + I+ ++++
Sbjct: 286 KKLKLAIWDTAGQERFRTLTSSYYRGAQGIIMVYDVTRRETFTNLSEIWAKEIDLYSTNQ 465
Query: 120 NVNKILVGNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDI-- 177
+ K+LVGNK D E R V +G A EYG F E SAKT +NV++ F + +
Sbjct: 466 DCMKMLVGNKVD-KEGDRVVTKKEGIDFAREYGCLFIECSAKTRVNVQQCFEELVLKVLD 642
Query: 178 KQRLADTDSKSEPQTIKINQPDQGAGAAQAAQKSACC 214
L SK + I ++P A A S CC
Sbjct: 643 TPSLLAEGSKGVKKNIFKDKPQSDASA------SGCC 735
>TC84871 similar to GP|1370172|emb|CAA98163.1 RAB1X {Lotus japonicus},
partial (87%)
Length = 727
Score = 166 bits (421), Expect = 5e-42
Identities = 91/172 (52%), Positives = 119/172 (68%), Gaps = 2/172 (1%)
Frame = +3
Query: 10 ADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIW 69
A YD K+LLIGDS VGKS LL+ F GS TIG+DFKI+ + + GKR+KL IW
Sbjct: 120 AAYDISFKILLIGDSAVGKSSLLVSFISGSVEDP-APTIGVDFKIKLLTVGGKRLKLTIW 296
Query: 70 DTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRN-WIRNIEQHASD-NVNKILVG 127
DTAGQERFRT+T++YYRGA GI+LVYDVT +F N+ W + +E ++++ + K+LVG
Sbjct: 297 DTAGQERFRTLTSSYYRGAQGIILVYDVTRRETFTNLSEVWSKEVELYSTNQDCMKMLVG 476
Query: 128 NKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQ 179
NK D ES+R V T +G ALA E+G FFE SAKT NV++ F +A I +
Sbjct: 477 NKVDR-ESERTVSTEEGLALAKEFGCSFFECSAKTRENVDKCFEELALKIME 629
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.134 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,736,126
Number of Sequences: 36976
Number of extensions: 61563
Number of successful extensions: 430
Number of sequences better than 10.0: 114
Number of HSP's better than 10.0 without gapping: 370
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 372
length of query: 216
length of database: 9,014,727
effective HSP length: 92
effective length of query: 124
effective length of database: 5,612,935
effective search space: 696003940
effective search space used: 696003940
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC125368.9


