

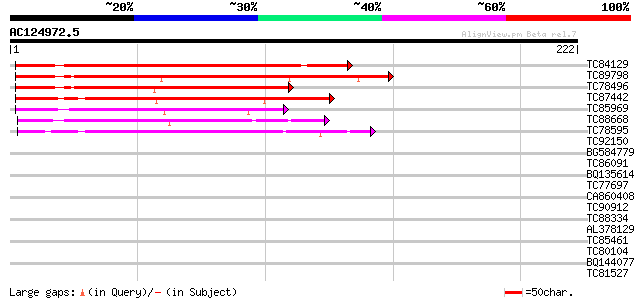
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124972.5 - phase: 0
(222 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC84129 weakly similar to PIR|T51492|T51492 hypothetical protein... 144 4e-35
TC89798 similar to PIR|T49936|T49936 hypothetical protein F17I14... 114 2e-26
TC78496 similar to PIR|T49936|T49936 hypothetical protein F17I14... 107 3e-24
TC87442 similar to GP|15809940|gb|AAL06897.1 AT4g05150/C17L7_70 ... 86 1e-17
TC85969 similar to GP|21553908|gb|AAM62991.1 unknown {Arabidopsi... 79 1e-15
TC88668 similar to GP|9293928|dbj|BAB01831.1 gb|AAF04880.1~gene_... 77 5e-15
TC78595 similar to PIR|T06718|T06718 hypothetical protein T29H11... 73 1e-13
TC92150 weakly similar to PIR|T47938|T47938 RING finger protein ... 31 0.43
BG584779 weakly similar to GP|15289962|db hypothetical protein~s... 30 0.57
TC86091 similar to PIR|T09896|T09896 hypothetical protein T22A6.... 30 0.57
BQ135614 similar to GP|13486729|dbj hypothetical protein {Oryza ... 30 0.74
TC77697 similar to GP|3335335|gb|AAC27137.1| ESTs gb|F14113 and ... 29 1.6
CA860408 GP|10728858|g CG10600 gene product {Drosophila melanoga... 28 2.1
TC90912 GP|7291633|gb|AAF47056.1| CG12491-PA {Drosophila melanog... 28 2.1
TC88334 similar to GP|13937204|gb|AAK50095.1 AT4g35220/F23E12_22... 28 2.1
AL378129 GP|20385063|g serine/threonine protein phosphatase 2A {... 28 2.1
TC85461 similar to GP|22324964|gb|AAM95691.1 hypothetical protei... 28 2.8
TC80104 similar to GP|20197699|gb|AAD20910.3 putative LRR recept... 28 2.8
BQ144077 weakly similar to GP|6523547|emb| hydroxyproline-rich g... 28 3.7
TC81527 similar to GP|9954116|gb|AAG08961.1| tuber-specific and ... 28 3.7
>TC84129 weakly similar to PIR|T51492|T51492 hypothetical protein T21H19_140
- Arabidopsis thaliana, partial (6%)
Length = 587
Score = 144 bits (362), Expect = 4e-35
Identities = 76/132 (57%), Positives = 95/132 (71%)
Frame = +1
Query: 3 RVKLMITYGGKIQPRLTALHDHLRYSYIGGDNKIITVDRNINFSDLMAKLSTFMFSDVCF 62
RVKLMI +GGKI RL HD YSYIGGD KI+TVDR+I FS L+ KLS+ FSD+C
Sbjct: 151 RVKLMIRFGGKIDLRL---HDDQNYSYIGGDTKILTVDRSIKFSSLIEKLSSMTFSDLCL 321
Query: 63 KYQLLGEDFDALIPIYNEEDLNHIMFEYDRMCHFSRKPAWLRVFLFPVPINNNKASFDSL 122
KYQ G + D+LI ++N++DL+ +MFEYD MC S KPA ++VFLFP+P+NN AS DSL
Sbjct: 322 KYQQPGGELDSLISVFNDDDLDSMMFEYDCMCRVSPKPARMKVFLFPLPVNN--ASSDSL 495
Query: 123 ASVKSLNFVRVS 134
S +L V S
Sbjct: 496 DSAFNLAVVEDS 531
>TC89798 similar to PIR|T49936|T49936 hypothetical protein F17I14.190 -
Arabidopsis thaliana, partial (19%)
Length = 1225
Score = 114 bits (286), Expect = 2e-26
Identities = 68/158 (43%), Positives = 96/158 (60%), Gaps = 10/158 (6%)
Frame = +3
Query: 3 RVKLMITYGGKIQPRLTALHDHLRYSYIGGDNKIITVDRNINFSDLMAKLSTFMFS-DVC 61
+ K M +YGGKIQPR HD+ + SY+GG+ KI+ VDRNI FS +++KLS+ + + DV
Sbjct: 165 KAKFMCSYGGKIQPRT---HDN-QLSYVGGETKILAVDRNIKFSSMISKLSSLIEAHDVS 332
Query: 62 FKYQLLGEDFDALIPIYNEEDLNHIMFEYDRMCHFSRKPAWLRVFLF-----PVPINNNK 116
FKYQL GED DALI + N++DL+H+M EYDR+ S +PA +R+FLF P P
Sbjct: 333 FKYQLPGEDLDALISVTNDDDLDHMMNEYDRLYRASARPARMRLFLFVNDSVPSPDPVKP 512
Query: 117 ASFDSLASVKSLNFVRVSP----FEEYSSPPTTPPPPF 150
++ D L ++ V P F + P P P +
Sbjct: 513 SNVDYLFGIEKPVTVAPIPAAVKFHDAVPEPVAPVPEY 626
>TC78496 similar to PIR|T49936|T49936 hypothetical protein F17I14.190 -
Arabidopsis thaliana, partial (23%)
Length = 1885
Score = 107 bits (268), Expect = 3e-24
Identities = 57/112 (50%), Positives = 75/112 (66%), Gaps = 3/112 (2%)
Frame = +1
Query: 3 RVKLMITYGGKIQPRLTALHDHLRYSYIGGDNKIITVDRNINFSDLMAKLSTF---MFSD 59
+ K M +YGGKIQPR HD+ + SYIGGD KI+ VDR+I F ++KLST + D
Sbjct: 433 KAKFMCSYGGKIQPRS---HDN-QLSYIGGDTKILAVDRSIKFQAFLSKLSTLCDALQQD 600
Query: 60 VCFKYQLLGEDFDALIPIYNEEDLNHIMFEYDRMCHFSRKPAWLRVFLFPVP 111
+ FKYQL GE+ DALI + E+DL H+M EYDR+ S KP +R+F+F P
Sbjct: 601 ISFKYQLPGEELDALISVTTEDDLEHMMHEYDRLYRPSSKPVRMRLFIFIAP 756
>TC87442 similar to GP|15809940|gb|AAL06897.1 AT4g05150/C17L7_70
{Arabidopsis thaliana}, partial (26%)
Length = 1572
Score = 85.5 bits (210), Expect = 1e-17
Identities = 53/127 (41%), Positives = 80/127 (62%), Gaps = 2/127 (1%)
Frame = +3
Query: 3 RVKLMITYGGKIQPRLTALHDHLRYSYIGGDNKIITVDRNINFSDLMAKLSTFM-FSDVC 61
RV+ M ++GGKI PR + + LRY +GGD +I+ V+R+I+FS L+ KLS S++
Sbjct: 165 RVRFMCSFGGKILPRPS--DNQLRY--VGGDTRIVAVNRSISFSALVHKLSKLSGMSNIT 332
Query: 62 FKYQLLGEDFDALIPIYNEEDLNHIMFEYDRMCHFSR-KPAWLRVFLFPVPINNNKASFD 120
KYQL ED DALI + +ED+++++ EYDR+ + A LR+FLFP ++ S
Sbjct: 333 AKYQLPNEDLDALITVTTDEDVDNMIDEYDRVTQSENPRAARLRLFLFPEGEDSRTNSIS 512
Query: 121 SLASVKS 127
SL + S
Sbjct: 513 SLLNGSS 533
>TC85969 similar to GP|21553908|gb|AAM62991.1 unknown {Arabidopsis
thaliana}, partial (31%)
Length = 2756
Score = 79.0 bits (193), Expect = 1e-15
Identities = 45/111 (40%), Positives = 67/111 (59%), Gaps = 4/111 (3%)
Frame = +2
Query: 3 RVKLMITYGGKIQPRLTALHDHLRYSYIGGDNKIITVDRNINFSDLMAKLSTFMFSD--V 60
+++LM +YGG I PR T Y+GGD +I+ V+R + DL +LS + +
Sbjct: 269 KLRLMCSYGGHIVPRPT----DKALCYVGGDTRIVVVERATSLGDLSTRLSKTLLNGRPF 436
Query: 61 CFKYQLLGEDFDALIPIYNEEDLNHIMFEYDR--MCHFSRKPAWLRVFLFP 109
FKYQL ED D+LI + +EDL +++ EYDR + S KP+ +R+FLFP
Sbjct: 437 TFKYQLPNEDLDSLISVSTDEDLENMIDEYDRTNSTNSSLKPSRIRLFLFP 589
>TC88668 similar to GP|9293928|dbj|BAB01831.1
gb|AAF04880.1~gene_id:MFE16.2~similar to unknown protein
{Arabidopsis thaliana}, partial (39%)
Length = 802
Score = 77.0 bits (188), Expect = 5e-15
Identities = 47/123 (38%), Positives = 74/123 (59%), Gaps = 1/123 (0%)
Frame = +2
Query: 4 VKLMITYGGKIQPRLTALHDHLRYSYIGGDNKIITVDRNINFSDLMAKLSTFMFSDVC-F 62
+K + +YGGKI PR + + Y+GG N++++VDR+I FS+L+ KL S V
Sbjct: 194 IKFLCSYGGKILPR----YPDGKLRYLGGHNRVLSVDRSIQFSELLLKLKELCSSSVTQL 361
Query: 63 KYQLLGEDFDALIPIYNEEDLNHIMFEYDRMCHFSRKPAWLRVFLFPVPINNNKASFDSL 122
+ QL ED DAL+ I ++EDL +++ EYDR + ++ F+ P P ++NKAS L
Sbjct: 362 RCQLPAEDLDALVSITSDEDLINLIEEYDRTAS-PQSQLKIKAFISP-PRSSNKASKPPL 535
Query: 123 ASV 125
S+
Sbjct: 536 PSL 544
>TC78595 similar to PIR|T06718|T06718 hypothetical protein T29H11.240 -
Arabidopsis thaliana, partial (46%)
Length = 1059
Score = 72.8 bits (177), Expect = 1e-13
Identities = 54/142 (38%), Positives = 78/142 (54%), Gaps = 2/142 (1%)
Frame = +1
Query: 4 VKLMITYGGKIQPRLTALHDHLRYSYIGGDNKIITVDRNINFSDLMAKLSTFMFSDVCFK 63
+K++ +YGGKI RL + LRY +GG +++ VDR+I+FS+LM KL S V +
Sbjct: 157 IKILYSYGGKI--RLRSTDGELRY--VGGHTRVLAVDRSISFSELMVKLEELCGSSVTLR 324
Query: 64 YQLLGEDFDALIPIYNEEDLNHIMFEYDRMCHFSRKPAWLRVFLFPVPINNNKASFD--S 121
QL D + LI I N+EDL +I+ EY+R P ++ L P ++ K S D S
Sbjct: 325 CQLPNGDLETLISITNDEDLTNIIDEYERASLKLTHPLKIKAIL-SQPKSSLKVSPDLSS 501
Query: 122 LASVKSLNFVRVSPFEEYSSPP 143
+S SL+ R SP S P
Sbjct: 502 SSSNASLSPAR-SPHTSAESLP 564
>TC92150 weakly similar to PIR|T47938|T47938 RING finger protein -
Arabidopsis thaliana, partial (51%)
Length = 595
Score = 30.8 bits (68), Expect = 0.43
Identities = 16/40 (40%), Positives = 22/40 (55%)
Frame = +1
Query: 118 SFDSLASVKSLNFVRVSPFEEYSSPPTTPPPPFMATKDTV 157
S S +SV S + +S SS PT+PPPP++ TV
Sbjct: 58 SLSSPSSVLSPSPSSLSSISPTSSTPTSPPPPYLTPTSTV 177
>BG584779 weakly similar to GP|15289962|db hypothetical protein~similar to
NBS-LRR type resistance protein, partial (4%)
Length = 782
Score = 30.4 bits (67), Expect = 0.57
Identities = 39/167 (23%), Positives = 63/167 (37%), Gaps = 26/167 (15%)
Frame = +1
Query: 3 RVKLMITYGGKIQPRLTA---LHDHLRYSYIGGDNKIITVDRNINFSDLMAKLSTFMFSD 59
RV L+ G K L + L D + Y Y DL+ L+ ++ D
Sbjct: 31 RVTLLKDIGNKFLKELRSRSFLQDFIDYGYAC----------KFKLHDLVHDLALYVSRD 180
Query: 60 VCFKYQLLGEDFDALIPIYNEEDLNHIMFEYDRMCHFSRKPAWLRVFLFPVPINN----- 114
++QLL D + E+ H+ F + + + P LR LFPV +NN
Sbjct: 181 ---EFQLLNSHSDNI-----SENALHLSFTKNYLFGKTPLPRGLRTILFPVGVNNEAFLN 336
Query: 115 --------------NKASFDSLAS----VKSLNFVRVSPFEEYSSPP 143
N + ++SL S +K L ++ + E+ S P
Sbjct: 337 TLVSRCTCLRVLQINNSGYESLPSSIGKLKHLRYLNLEDNEKLKSVP 477
>TC86091 similar to PIR|T09896|T09896 hypothetical protein T22A6.160 -
Arabidopsis thaliana, partial (69%)
Length = 1865
Score = 30.4 bits (67), Expect = 0.57
Identities = 22/87 (25%), Positives = 36/87 (41%), Gaps = 10/87 (11%)
Frame = +2
Query: 123 ASVKSLNFVRVSPFEE-YSSPPTTPPPPFMATKDTVTKAEMKKFRGLHVVNDEFEDILKL 181
+ V LN +S F+ +PP TP PP + K L+ V ++
Sbjct: 368 SKVFPLNHTLISKFQPPIPNPPITPTPPLLIIKTL----------SLNHVPSPLRSFAEV 517
Query: 182 YLTCNFS---------KVKDFHGLHVV 199
+L+C+ S + FHGLH++
Sbjct: 518 FLSCSLSITSLVKRKTRT*HFHGLHIL 598
>BQ135614 similar to GP|13486729|dbj hypothetical protein {Oryza sativa
(japonica cultivar-group)}, partial (11%)
Length = 1384
Score = 30.0 bits (66), Expect = 0.74
Identities = 17/37 (45%), Positives = 19/37 (50%)
Frame = -1
Query: 136 FEEYSSPPTTPPPPFMATKDTVTKAEMKKFRGLHVVN 172
F SP TTPPPPF T T K F GL V++
Sbjct: 607 FRSIPSPITTPPPPFRPT----THPRRKLFPGLVVLS 509
>TC77697 similar to GP|3335335|gb|AAC27137.1| ESTs gb|F14113 and gb|T42122
come from this region. {Arabidopsis thaliana}, partial
(78%)
Length = 1365
Score = 28.9 bits (63), Expect = 1.6
Identities = 12/36 (33%), Positives = 17/36 (46%)
Frame = +2
Query: 114 NNKASFDSLASVKSLNFVRVSPFEEYSSPPTTPPPP 149
+N SF S + V + P + +PP PPPP
Sbjct: 206 SNPTSFSSTMKNSASKAVALKPVHRHLTPPPPPPPP 313
>CA860408 GP|10728858|g CG10600 gene product {Drosophila melanogaster},
partial (0%)
Length = 451
Score = 28.5 bits (62), Expect = 2.1
Identities = 14/43 (32%), Positives = 21/43 (48%)
Frame = +2
Query: 119 FDSLASVKSLNFVRVSPFEEYSSPPTTPPPPFMATKDTVTKAE 161
F SL+S+ S + + S+PP PPPPF + + E
Sbjct: 122 FSSLSSLWSSSSISRHSSTRTSAPPPPPPPPFKPDQSVLLVGE 250
>TC90912 GP|7291633|gb|AAF47056.1| CG12491-PA {Drosophila melanogaster},
partial (7%)
Length = 524
Score = 28.5 bits (62), Expect = 2.1
Identities = 23/60 (38%), Positives = 31/60 (51%)
Frame = +3
Query: 103 LRVFLFPVPINNNKASFDSLASVKSLNFVRVSPFEEYSSPPTTPPPPFMATKDTVTKAEM 162
LR F P P +N +S DS +S S+ V S S+P +TPPPP + TK E+
Sbjct: 252 LRQFSPPQP--SNDSSSDSSSSTPSV--VVFSDQPSSSAPDSTPPPP--SPSPATTKIEV 413
>TC88334 similar to GP|13937204|gb|AAK50095.1 AT4g35220/F23E12_220
{Arabidopsis thaliana}, partial (83%)
Length = 1314
Score = 28.5 bits (62), Expect = 2.1
Identities = 19/57 (33%), Positives = 23/57 (40%), Gaps = 5/57 (8%)
Frame = +1
Query: 136 FEEYSSPPTTPPPPFMATKDTVTKAEMKKFRGLHVVNDEFEDILKLY-----LTCNF 187
F PP PP P + T TV MK F LH V D ++ L+C F
Sbjct: 187 FPSPQPPPLIPPSPAL-TPATVASPAMKLFSFLHDVKSTMPDEYSIFPTSTPLSCRF 354
>AL378129 GP|20385063|g serine/threonine protein phosphatase 2A {Pisum
sativum}, partial (37%)
Length = 421
Score = 28.5 bits (62), Expect = 2.1
Identities = 15/43 (34%), Positives = 22/43 (50%)
Frame = -1
Query: 107 LFPVPINNNKASFDSLASVKSLNFVRVSPFEEYSSPPTTPPPP 149
LF V K F+ + ++ +L V +P + SPP PPPP
Sbjct: 148 LFNVFAEKLKFVFEKMLAIFNLRDVSGTPQKTRRSPPYPPPPP 20
>TC85461 similar to GP|22324964|gb|AAM95691.1 hypothetical protein {Oryza
sativa (japonica cultivar-group)}, partial (52%)
Length = 1376
Score = 28.1 bits (61), Expect = 2.8
Identities = 16/33 (48%), Positives = 19/33 (57%), Gaps = 1/33 (3%)
Frame = -2
Query: 118 SFDSLASVKSLNFVRVS-PFEEYSSPPTTPPPP 149
SF SL ++ SLN V F + SPP PPPP
Sbjct: 844 SFFSLFNLSSLNSSNVFFLFSPFLSPPPFPPPP 746
>TC80104 similar to GP|20197699|gb|AAD20910.3 putative LRR receptor protein
kinase {Arabidopsis thaliana}, partial (28%)
Length = 1549
Score = 28.1 bits (61), Expect = 2.8
Identities = 17/50 (34%), Positives = 27/50 (54%), Gaps = 4/50 (8%)
Frame = +1
Query: 104 RVFLFPVPINNNKASFD--SLASVKSL--NFVRVSPFEEYSSPPTTPPPP 149
RV+ P+P ++K D +A++ + + +S E YS PP PPPP
Sbjct: 835 RVWAVPIPNAHDKQEKDVQRMATIAKPVDHEIDISTPEVYSVPPPPPPPP 984
>BQ144077 weakly similar to GP|6523547|emb| hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (16%)
Length = 463
Score = 27.7 bits (60), Expect = 3.7
Identities = 17/48 (35%), Positives = 20/48 (41%)
Frame = +3
Query: 107 LFPVPINNNKASFDSLASVKSLNFVRVSPFEEYSSPPTTPPPPFMATK 154
LFP P+ F L F F E SPP +PPPP A +
Sbjct: 138 LFPFPL------FPCLPFPSPPGFPPPPHFPETPSPPPSPPPPSYANR 263
>TC81527 similar to GP|9954116|gb|AAG08961.1| tuber-specific and
sucrose-responsive element binding factor {Solanum
tuberosum}, partial (56%)
Length = 765
Score = 27.7 bits (60), Expect = 3.7
Identities = 20/75 (26%), Positives = 30/75 (39%), Gaps = 8/75 (10%)
Frame = +2
Query: 135 PFEEYSSPPTTPPPPFMATKDTVTKAEMKKFRGLHVVNDEFEDILK--------LYLTCN 186
P E+ T PP +D +F V EF D+++ Y++ N
Sbjct: 521 PLEDDPLTALTLAPPGNGVEDREEMVGDHRFPSPENVPTEFWDVMRGVIAREVREYVSSN 700
Query: 187 FSKVKDFHGLHVVND 201
FS FH + +VND
Sbjct: 701 FSHNSSFH*IELVND 745
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.141 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,039,858
Number of Sequences: 36976
Number of extensions: 129429
Number of successful extensions: 2751
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 1676
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2671
length of query: 222
length of database: 9,014,727
effective HSP length: 92
effective length of query: 130
effective length of database: 5,612,935
effective search space: 729681550
effective search space used: 729681550
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC124972.5


