

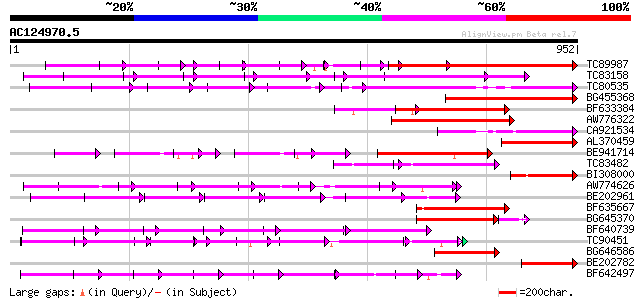
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124970.5 - phase: 0
(952 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC89987 weakly similar to PIR|D86443|D86443 probable PPR-repeat ... 288 4e-91
TC83158 weakly similar to PIR|H86191|H86191 hypothetical protein... 256 2e-68
TC80535 weakly similar to GP|14335136|gb|AAK59848.1 At2g15690/F9... 233 2e-61
BG455368 weakly similar to GP|9759524|dbj selenium-binding prote... 202 5e-52
BF633384 similar to PIR|T46179|T46 hypothetical protein T8H10.30... 162 7e-40
AW776322 weakly similar to PIR|H84859|H84 hypothetical protein A... 158 1e-38
CA921534 similar to PIR|D96516|D96 F16N3.14 [imported] - Arabido... 157 2e-38
AL370459 weakly similar to GP|20146256|d hypothetical protein {O... 156 4e-38
BE941714 weakly similar to GP|11994734|dbj selenium-binding prot... 150 3e-36
TC83482 weakly similar to GP|9758453|dbj|BAB08982.1 selenium-bin... 139 4e-33
BI308000 similar to GP|7363286|dbj| Similar to Arabidopsis thali... 133 3e-31
AW774626 similar to PIR|T47909|T47 hypothetical protein T20K12.7... 131 1e-30
BE202961 weakly similar to GP|6016735|gb|A hypothetical protein ... 130 3e-30
BF635667 weakly similar to PIR|C85359|C853 hypothetical protein ... 129 5e-30
BG645370 weakly similar to PIR|T00405|T004 hypothetical protein ... 122 1e-29
BF640739 weakly similar to GP|7363286|dbj Similar to Arabidopsis... 119 4e-27
TC90451 similar to PIR|H84508|H84508 hypothetical protein At2g13... 118 1e-26
BG646586 weakly similar to GP|10177683|dbj selenium-binding prot... 108 1e-23
BE202782 weakly similar to GP|22093801|dbj hypothetical protein~... 108 1e-23
BF642497 weakly similar to PIR|H96713|H96 hypothetical protein T... 107 3e-23
>TC89987 weakly similar to PIR|D86443|D86443 probable PPR-repeat protein
[imported] - Arabidopsis thaliana, partial (62%)
Length = 1559
Score = 288 bits (737), Expect(2) = 4e-91
Identities = 136/316 (43%), Positives = 202/316 (63%)
Frame = +2
Query: 637 VFDMMPIKDVFSWNTMIFANGMHGNGKEALSLFEKMLLSMVKPDSATFTCVLSACSHSML 696
VF M K+ +S+ MI +HG GKEAL +F +M+ + PD + V SACSH+ L
Sbjct: 581 VFKNMSEKNRYSYTVMISGLAIHGRGKEALKVFSEMIEEGLAPDDVVYVGVFSACSHAGL 760
Query: 697 VEEGVQIFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAYGFIQRMPMEPTAIAWKAFL 756
VEEG+Q F SM +H +EP +HY C+VD+ R G L+EAY I+ M ++P + W++ L
Sbjct: 761 VEEGLQCFKSMQFEHKIEPTVQHYGCMVDLLGRFGMLKEAYELIKSMSIKPNDVIWRSLL 940
Query: 757 AGCRVYKNVELAKISAKKLFEIDPNGSANYVTLFNILVTAKLWSEASKIRKLMKERGITK 816
+ C+V+ N+E+ KI+A+ LF ++ N S +Y+ L N+ A+ W + +KIR + ER + +
Sbjct: 941 SACKVHHNLEIGKIAAENLFMLNQNNSGDYLVLANMYAKAQKWDDVAKIRTKLAERNLVQ 1120
Query: 817 TPGCSWFHVGNRVHTFVAGDKSNMESDKIYNFLDELFAKIKAAGYKPDTDYVLHDIDQEE 876
TPG S +V+ FV+ DKS + + IY + ++ ++K GY PDT VL D+D EE
Sbjct: 1121TPGFSLIEAKRKVYKFVSQDKSIPQWNIIYEMIHQMEWQVKFEGYIPDTSQVLLDVDDEE 1300
Query: 877 KAESLCNHSEKLAVAFGILNLNGQSTIRVFKNLRICGDCHNAIKYMSNVVGVTIVVRDSL 936
K E L HS+KLA+AFG+++ + +R+ +NLR+C DCH KY+S + I VRD
Sbjct: 1301KKERLKFHSQKLAIAFGLIHTSEGXPLRITRNLRMCSDCHTYTKYISMIYEREITVRDRX 1480
Query: 937 RFHHFKNGNCSCKDFW 952
RF HFKNG+CSCKD+W
Sbjct: 1481RFXHFKNGSCSCKDYW 1528
Score = 76.3 bits (186), Expect = 5e-14
Identities = 50/155 (32%), Positives = 83/155 (53%), Gaps = 1/155 (0%)
Frame = +1
Query: 589 ACSLSECLRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIKDVFS 648
ACSL + G ++H +VF+ + D+ N+L++MY KCG + + +VF+ M K V S
Sbjct: 130 ACSLLGVVDEGIQVHGHVFKMGLEGDVIVQNSLINMYGKCGEIKNACDVFNGMDEKSVAS 309
Query: 649 WNTMIFANGMHGNGKEALSLFEKMLL-SMVKPDSATFTCVLSACSHSMLVEEGVQIFNSM 707
W+ +I A+ E L L KM + + +T VLSAC+H + G I +
Sbjct: 310 WSAIIGAHACVEMWNECLMLLGKMSSEGRCRVEESTLVNVLSACTHLGSPDLGKCIHGIL 489
Query: 708 SRDHLVEPEAEHYTCVVDIYSRAGCLEEAYGFIQR 742
R ++ E T ++D+Y ++GCLE+ + IQ+
Sbjct: 490 LR-NISELNVVVKTSLIDMYVKSGCLEKRFTRIQK 591
Score = 73.2 bits (178), Expect = 5e-13
Identities = 42/146 (28%), Positives = 76/146 (51%), Gaps = 1/146 (0%)
Frame = +1
Query: 151 CSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVTW 210
CS L + G ++HG V + G+ DV V ++ +N Y KC ++ A VF+ M + V +W
Sbjct: 133 CSLLGVVDEGIQVHGHVFKMGLEGDVIVQNSLINMYGKCGEIKNACDVFNGMDEKSVASW 312
Query: 211 NSLSSCYVNCGFPQKGLNVFREMVLDG-VKPDPVTVSCILSACSDLQDLKSGKAIHGFAL 269
+++ + + L + +M +G + + T+ +LSAC+ L GK IHG L
Sbjct: 313 SAIIGAHACVEMWNECLMLLGKMSSEGRCRVEESTLVNVLSACTHLGSPDLGKCIHGILL 492
Query: 270 KHGMVENVFVSNALVNLYESCLCVRE 295
++ NV V +L+++Y C+ +
Sbjct: 493 RNISELNVVVKTSLIDMYVKSGCLEK 570
Score = 71.6 bits (174), Expect(2) = 7e-21
Identities = 42/147 (28%), Positives = 76/147 (51%), Gaps = 1/147 (0%)
Frame = +1
Query: 352 ACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMPHRNVVT 411
ACS L + G +HG K G+ DV V +L+N+Y C ++ A VF+ M ++V +
Sbjct: 130 ACSLLGVVDEGIQVHGHVFKMGLEGDVIVQNSLINMYGKCGEIKNACDVFNGMDEKSVAS 309
Query: 412 WNSLSSCYVNCGFPQKGLNVFREMVLNG-VKPDLVTMLSILHACSDLQDLKSGKVIHGFA 470
W+++ + + L + +M G + + T++++L AC+ L GK IHG
Sbjct: 310 WSAIIGAHACVEMWNECLMLLGKMSSEGRCRVEESTLVNVLSACTHLGSPDLGKCIHGIL 489
Query: 471 VRHGMVEDVFVCNALLSLYAKCVCVRE 497
+R+ +V V +L+ +Y K C+ +
Sbjct: 490 LRNISELNVVVKTSLIDMYVKSGCLEK 570
Score = 70.5 bits (171), Expect(2) = 2e-17
Identities = 41/150 (27%), Positives = 77/150 (51%), Gaps = 1/150 (0%)
Frame = +1
Query: 251 ACSDLQDLKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVIT 310
ACS L + G +HG K G+ +V V N+L+N+Y C ++ A VF+ M ++V +
Sbjct: 130 ACSLLGVVDEGIQVHGHVFKMGLEGDVIVQNSLINMYGKCGEIKNACDVFNGMDEKSVAS 309
Query: 311 WNSLASCYVNCGFPQKGLNVFREMGLNG-VKPDPMAMSSILPACSQLKDLKSGKTIHGFA 369
W+++ + + L + +M G + + + ++L AC+ L GK IHG
Sbjct: 310 WSAIIGAHACVEMWNECLMLLGKMSSEGRCRVEESTLVNVLSACTHLGSPDLGKCIHGIL 489
Query: 370 VKHGMVEDVFVCTALVNLYANCLCVREAQT 399
+++ +V V T+L+++Y C+ + T
Sbjct: 490 LRNISELNVVVKTSLIDMYVKSGCLEKRFT 579
Score = 66.2 bits (160), Expect(2) = 4e-91
Identities = 46/179 (25%), Positives = 84/179 (46%)
Frame = +1
Query: 453 ACSDLQDLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHREVAS 512
ACS L + G +HG + G+ DV V N+L+++Y KC ++ A VF+ + + VAS
Sbjct: 130 ACSLLGVVDEGIQVHGHVFKMGLEGDVIVQNSLINMYGKCGEIKNACDVFNGMDEKSVAS 309
Query: 513 WNGILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGGCVKNSRIEEAMEIFRKMQ 572
W+ I+ A+ + + + L + +M+ + G C
Sbjct: 310 WSAIIGAHACVEMWNECLMLLGKMSSE-------------GRC----------------- 399
Query: 573 TMGFKPDETTIYSILRACSLSECLRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCGGL 631
+ +E+T+ ++L AC+ +GK IH + R+ + ++ +L+DMY K G L
Sbjct: 400 ----RVEESTLVNVLSACTHLGSPDLGKCIHGILLRNISELNVVVKTSLIDMYVKSGCL 564
Score = 60.8 bits (146), Expect(2) = 4e-15
Identities = 35/138 (25%), Positives = 68/138 (48%), Gaps = 1/138 (0%)
Frame = +1
Query: 61 QFHDDATRCGVMSDVSIGNAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCG 120
Q H + G+ DV + N+ I+ YGKC ++ A VF+ + + V +W+++ +
Sbjct: 166 QVHGHVFKMGLEGDVIVQNSLINMYGKCGEIKNACDVFNGMDEKSVASWSAIIGAHACVE 345
Query: 121 FPQQGLNVFRKMGL-NKVKANPLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVS 179
+ L + KM + + T+ ++L C+ L GK IHG ++R+ +V V
Sbjct: 346 MWNECLMLLGKMSSEGRCRVEESTLVNVLSACTHLGSPDLGKCIHGILLRNISELNVVVK 525
Query: 180 SAFVNFYAKCLCVREAQT 197
++ ++ Y K C+ + T
Sbjct: 526 TSLIDMYVKSGCLEKRFT 579
Score = 48.1 bits (113), Expect(2) = 7e-21
Identities = 36/139 (25%), Positives = 67/139 (47%), Gaps = 5/139 (3%)
Frame = +2
Query: 528 KGLYMFSQMNRDEVKADEITWSVVIGGCVKNSRIEEAMEIFRKMQTMGFKPDETTIYSIL 587
KGL +F M+ + + +++V+I G + R +EA+++F +M G PD+ +
Sbjct: 569 KGLRVFKNMS----EKNRYSYTVMISGLAIHGRGKEALKVFSEMIEEGLAPDDVVYVGVF 736
Query: 588 RACSLSECLRMGKEIHCY---VFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIK 644
ACS + + G + C+ F H + + +VD+ + G L + + M IK
Sbjct: 737 SACSHAGLVEEG--LQCFKSMQFEHKIEPTVQHYGCMVDLLGRFGMLKEAYELIKSMSIK 910
Query: 645 --DVFSWNTMIFANGMHGN 661
DV W +++ A +H N
Sbjct: 911 PNDVI-WRSLLSACKVHHN 964
Score = 39.3 bits (90), Expect(2) = 4e-15
Identities = 32/127 (25%), Positives = 57/127 (44%), Gaps = 2/127 (1%)
Frame = +2
Query: 192 VREAQTVFDLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSA 251
+R+ VF M ++ ++ + S G ++ L VF EM+ +G+ PD V + SA
Sbjct: 563 LRKGLRVFKNMSEKNRYSYTVMISGLAIHGRGKEALKVFSEMIEEGLAPDDVVYVGVFSA 742
Query: 252 CSDLQDLKSG-KAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHR-NVI 309
CS ++ G + +H + V +V+L ++EA + M + N +
Sbjct: 743 CSHAGLVEEGLQCFKSMQFEHKIEPTVQHYGCMVDLLGRFGMLKEAYELIKSMSIKPNDV 922
Query: 310 TWNSLAS 316
W SL S
Sbjct: 923 IWRSLLS 943
Score = 37.7 bits (86), Expect(2) = 2e-17
Identities = 36/155 (23%), Positives = 65/155 (41%), Gaps = 11/155 (7%)
Frame = +2
Query: 394 VREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHA 453
+R+ VF M +N ++ + S G ++ L VF EM+ G+ PD V + + A
Sbjct: 563 LRKGLRVFKNMSEKNRYSYTVMISGLAIHGRGKEALKVFSEMIEEGLAPDDVVYVGVFSA 742
Query: 454 CSDLQDLKSG-KVIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHREV-- 510
CS ++ G + H + V ++ L + ++EA ++LI +
Sbjct: 743 CSHAGLVEEGLQCFKSMQFEHKIEPTVQHYGCMVDLLGRFGMLKEA---YELIKSMSIKP 913
Query: 511 --ASWNGILTAYFTNKEYEKG------LYMFSQMN 537
W +L+A + E G L+M +Q N
Sbjct: 914 NDVIWRSLLSACKVHHNLEIGKIAAENLFMLNQNN 1018
Score = 33.9 bits (76), Expect = 0.30
Identities = 26/139 (18%), Positives = 54/139 (38%), Gaps = 33/139 (23%)
Frame = +2
Query: 495 VREAQVVFDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGG 554
+R+ VF + + S+ +++ + ++ L +FS+M + + D++ + V
Sbjct: 563 LRKGLRVFKNMSEKNRYSYTVMISGLAIHGRGKEALKVFSEMIEEGLAPDDVVYVGVFSA 742
Query: 555 CVKNSRIEEAMEIFRKMQ---------------------------------TMGFKPDET 581
C +EE ++ F+ MQ +M KP++
Sbjct: 743 CSHAGLVEEGLQCFKSMQFEHKIEPTVQHYGCMVDLLGRFGMLKEAYELIKSMSIKPNDV 922
Query: 582 TIYSILRACSLSECLRMGK 600
S+L AC + L +GK
Sbjct: 923 IWRSLLSACKVHHNLEIGK 979
>TC83158 weakly similar to PIR|H86191|H86191 hypothetical protein [imported]
- Arabidopsis thaliana, partial (42%)
Length = 1194
Score = 256 bits (655), Expect = 2e-68
Identities = 133/328 (40%), Positives = 198/328 (59%)
Frame = +2
Query: 546 ITWSVVIGGCVKNSRIEEAMEIFRKMQTMGFKPDETTIYSILRACSLSECLRMGKEIHCY 605
++W+VVIGG VK EEA+E FR+MQ G PD T+ +I+ AC+ L +G +H
Sbjct: 56 VSWTVVIGGFVKKECYEEALECFREMQLAGVVPDFVTVIAIISACANLGALGLGLWVHRL 235
Query: 606 VFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGKEA 665
V + ++ N+L+DMYA+CG + L+R VFD M +++ SWN++I ++G +A
Sbjct: 236 VMKKEFRDNVKVLNSLIDMYARCGCIELARQVFDGMSQRNLVSWNSIIVGFAVNGLADKA 415
Query: 666 LSLFEKMLLSMVKPDSATFTCVLSACSHSMLVEEGVQIFNSMSRDHLVEPEAEHYTCVVD 725
LS F M ++P+ ++T L+ACSH+ L++EG++IF + RDH P EHY C+VD
Sbjct: 416 LSFFRSMKKEGLEPNGVSYTSALTACSHAGLIDEGLKIFADIKRDHRNSPRIEHYGCLVD 595
Query: 726 IYSRAGCLEEAYGFIQRMPMEPTAIAWKAFLAGCRVYKNVELAKISAKKLFEIDPNGSAN 785
+YSRAG L+EA+ I++MPM P + + LA CR +VELA+ K E+ P G +N
Sbjct: 596 LYSRAGRLKEAWDVIKKMPMMPNEVVLGSLLAACRTQGDVELAEKVMKYQVELYPGGDSN 775
Query: 786 YVTLFNILVTAKLWSEASKIRKLMKERGITKTPGCSWFHVGNRVHTFVAGDKSNMESDKI 845
YV NI W ASK+R+ MKERG+ K S + + +H FV+GDK + E+D I
Sbjct: 776 YVLFSNIYAAVGKWDGASKVRREMKERGLQKNLAFSSIEIDSGIHKFVSGDKYHEENDYI 955
Query: 846 YNFLDELFAKIKAAGYKPDTDYVLHDID 873
Y+ L+ L ++ GY PD D+D
Sbjct: 956 YSALELLSFELHLYGYVPDFSGKESDVD 1039
Score = 122 bits (305), Expect = 9e-28
Identities = 78/230 (33%), Positives = 121/230 (51%), Gaps = 3/230 (1%)
Frame = +2
Query: 192 VREAQTVFDLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSA 251
V +A +FD +P ++VV+W + +V ++ L FREM L GV PD VTV I+SA
Sbjct: 8 VDDALKLFDKLPVKNVVSWTVVIGGFVKKECYEEALECFREMQLAGVVPDFVTVIAIISA 187
Query: 252 CSDLQDLKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITW 311
C++L L G +H +K +NV V N+L+++Y C C+ A+ VFD M RN+++W
Sbjct: 188 CANLGALGLGLWVHRLVMKKEFRDNVKVLNSLIDMYARCGCIELARQVFDGMSQRNLVSW 367
Query: 312 NSLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSG-KTIHGFAV 370
NS+ + G K L+ FR M G++P+ ++ +S L ACS + G K
Sbjct: 368 NSIIVGFAVNGLADKALSFFRSMKKEGLEPNGVSYTSALTACSHAGLIDEGLKIFADIKR 547
Query: 371 KHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMP--HRNVVTWNSLSSC 418
H + LV+LY+ ++EA V MP VV + L++C
Sbjct: 548 DHRNSPRIEHYGCLVDLYSRAGRLKEAWDVIKKMPMMPNEVVLGSLLAAC 697
Score = 112 bits (281), Expect = 5e-25
Identities = 69/215 (32%), Positives = 112/215 (52%), Gaps = 1/215 (0%)
Frame = +2
Query: 293 VREAQAVFDLMPHRNVITWNSLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPA 352
V +A +FD +P +NV++W + +V ++ L FREM L GV PD + + +I+ A
Sbjct: 8 VDDALKLFDKLPVKNVVSWTVVIGGFVKKECYEEALECFREMQLAGVVPDFVTVIAIISA 187
Query: 353 CSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMPHRNVVTW 412
C+ L L G +H +K ++V V +L+++YA C C+ A+ VFD M RN+V+W
Sbjct: 188 CANLGALGLGLWVHRLVMKKEFRDNVKVLNSLIDMYARCGCIELARQVFDGMSQRNLVSW 367
Query: 413 NSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHACSDLQDLKSG-KVIHGFAV 471
NS+ + G K L+ FR M G++P+ V+ S L ACS + G K+
Sbjct: 368 NSIIVGFAVNGLADKALSFFRSMKKEGLEPNGVSYTSALTACSHAGLIDEGLKIFADIKR 547
Query: 472 RHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIP 506
H + L+ LY++ ++EA V +P
Sbjct: 548 DHRNSPRIEHYGCLVDLYSRAGRLKEAWDVIKKMP 652
Score = 112 bits (279), Expect = 9e-25
Identities = 56/175 (32%), Positives = 100/175 (57%)
Frame = +2
Query: 394 VREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHA 453
V +A +FD +P +NVV+W + +V ++ L FREM L GV PD VT+++I+ A
Sbjct: 8 VDDALKLFDKLPVKNVVSWTVVIGGFVKKECYEEALECFREMQLAGVVPDFVTVIAIISA 187
Query: 454 CSDLQDLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHREVASW 513
C++L L G +H ++ ++V V N+L+ +YA+C C+ A+ VFD + R + SW
Sbjct: 188 CANLGALGLGLWVHRLVMKKEFRDNVKVLNSLIDMYARCGCIELARQVFDGMSQRNLVSW 367
Query: 514 NGILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGGCVKNSRIEEAMEIF 568
N I+ + N +K L F M ++ ++ + ++++ + C I+E ++IF
Sbjct: 368 NSIIVGFAVNGLADKALSFFRSMKKEGLEPNGVSYTSALTACSHAGLIDEGLKIF 532
Score = 105 bits (263), Expect = 6e-23
Identities = 69/230 (30%), Positives = 119/230 (51%), Gaps = 3/230 (1%)
Frame = +2
Query: 91 VEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVKANPLTVSSILPG 150
V+ A ++FD L ++VV+W + +V ++ L FR+M L V + +TV +I+
Sbjct: 8 VDDALKLFDKLPVKNVVSWTVVIGGFVKKECYEEALECFREMQLAGVVPDFVTVIAIISA 187
Query: 151 CSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVTW 210
C++L L G +H V++ ++V V ++ ++ YA+C C+ A+ VFD M R++V+W
Sbjct: 188 CANLGALGLGLWVHRLVMKKEFRDNVKVLNSLIDMYARCGCIELARQVFDGMSQRNLVSW 367
Query: 211 NSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQDLKSG-KAIHGFAL 269
NS+ + G K L+ FR M +G++P+ V+ + L+ACS + G K
Sbjct: 368 NSIIVGFAVNGLADKALSFFRSMKKEGLEPNGVSYTSALTACSHAGLIDEGLKIFADIKR 547
Query: 270 KHGMVENVFVSNALVNLYESCLCVREAQAVFDLMP--HRNVITWNSLASC 317
H + LV+LY ++EA V MP V+ + LA+C
Sbjct: 548 DHRNSPRIEHYGCLVDLYSRAGRLKEAWDVIKKMPMMPNEVVLGSLLAAC 697
Score = 75.5 bits (184), Expect = 9e-14
Identities = 54/198 (27%), Positives = 97/198 (48%), Gaps = 4/198 (2%)
Frame = +2
Query: 23 EAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQF-HDDATRCGVMSDVSIGNAF 81
EA++ + + G+ PD +A+ ACA + AL + + H + +V + N+
Sbjct: 107 EALECFREMQLAGVVPDFVTVIAIISACA-NLGALGLGLWVHRLVMKKEFRDNVKVLNSL 283
Query: 82 IHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVKANP 141
I Y +C C+E AR+VFD + R++V+WNS+ + G + L+ FR M ++ N
Sbjct: 284 IDMYARCGCIELARQVFDGMSQRNLVSWNSIIVGFAVNGLADKALSFFRSMKKEGLEPNG 463
Query: 142 LTVSSILPGCSDLQDLKSGKEIHGFVVR-HGMVEDVFVSSAFVNFYAKCLCVREAQTVFD 200
++ +S L CS + G +I + R H + V+ Y++ ++EA V
Sbjct: 464 VSYTSALTACSHAGLIDEGLKIFADIKRDHRNSPRIEHYGCLVDLYSRAGRLKEAWDVIK 643
Query: 201 LMP--HRDVVTWNSLSSC 216
MP +VV + L++C
Sbjct: 644 KMPMMPNEVVLGSLLAAC 697
Score = 60.5 bits (145), Expect = 3e-09
Identities = 45/179 (25%), Positives = 82/179 (45%), Gaps = 2/179 (1%)
Frame = +2
Query: 629 GGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGKEALSLFEKMLLSMVKPDSATFTCVL 688
G + + +FD +P+K+V SW +I +EAL F +M L+ V PD T ++
Sbjct: 2 GDVDDALKLFDKLPVKNVVSWTVVIGGFVKKECYEEALECFREMQLAGVVPDFVTVIAII 181
Query: 689 SACSHSMLVEEGVQIFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAYGFIQRMPMEPT 748
SAC++ + G+ + + + + ++D+Y+R GC+E A M +
Sbjct: 182 SACANLGALGLGLWVHRLVMKKEF-RDNVKVLNSLIDMYARCGCIELARQVFDGM-SQRN 355
Query: 749 AIAWKAFLAGCRV--YKNVELAKISAKKLFEIDPNGSANYVTLFNILVTAKLWSEASKI 805
++W + + G V + L+ + K ++PNG +Y + A L E KI
Sbjct: 356 LVSWNSIIVGFAVNGLADKALSFFRSMKKEGLEPNG-VSYTSALTACSHAGLIDEGLKI 529
>TC80535 weakly similar to GP|14335136|gb|AAK59848.1 At2g15690/F9O13.24
{Arabidopsis thaliana}, partial (55%)
Length = 1727
Score = 233 bits (595), Expect = 2e-61
Identities = 134/398 (33%), Positives = 212/398 (52%), Gaps = 2/398 (0%)
Frame = +2
Query: 557 KNSRIEEAMEIFRKMQTMGFKPDETTIYSILRACSLSECLRMGKEIHCYVFRHWKDWDLA 616
+ +++EA+E+ K G K D + C S+ + K++H Y + D
Sbjct: 224 QEGKVKEALELMEK----GIKADANCFEILFDLCGKSKSVEDAKKVHDYFLQSTFRSDFK 391
Query: 617 RTNALVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGKEALSLFEKMLLSM 676
N +++MY C ++ +R VFD MP +++ SW+ MI G E L LFE+M
Sbjct: 392 MHNKVIEMYGNCKSMTDARRVFDHMPNRNMDSWHMMIRGYANSTMGDEGLQLFEQMNELG 571
Query: 677 VKPDSATFTCVLSACSHSMLVEEGVQIFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEA 736
++ S T VLSAC + VE+ SM + +EP EHY ++D+ ++G L+EA
Sbjct: 572 LEITSETMLAVLSACGSAEAVEDAYIYLESMKSKYGIEPGVEHYMGLLDVLGQSGYLKEA 751
Query: 737 YGFIQRMPMEPTAIAWKAFLAGCRVYKNVELAKISAKKLFEIDPNGSANYVTLFNILVTA 796
FI+++P EPT ++ R++ +V+L + + +DP+ + V
Sbjct: 752 EEFIEQLPFEPTVTVFETLKNYARIHGDVDLEDHVEELIVSLDPSKA----------VAN 901
Query: 797 KLWSEASKIRKLMKERGITKTPGCSWFHVGNRVHTFVAGDKSNMESDKIYNFLDELFA-- 854
K+ + K K S NR+ + ++ +Y ++L A
Sbjct: 902 KIPTPPPK-----------KYTAISMLDGKNRIIEY--------KNPTLYKDDEKLIAMN 1024
Query: 855 KIKAAGYKPDTDYVLHDIDQEEKAESLCNHSEKLAVAFGILNLNGQSTIRVFKNLRICGD 914
+K AGY PDT YVLHDIDQE K ++L HSE+LA+A+G+++ ++ +R+ KNLR+CGD
Sbjct: 1025SMKDAGYVPDTRYVLHDIDQEAKEQALLYHSERLAIAYGLISTPPRTPLRIIKNLRVCGD 1204
Query: 915 CHNAIKYMSNVVGVTIVVRDSLRFHHFKNGNCSCKDFW 952
CHNAIK MS +VG ++VRD+ RFHHFK+G CSC D+W
Sbjct: 1205CHNAIKIMSRIVGRELIVRDNKRFHHFKDGKCSCGDYW 1318
Score = 76.6 bits (187), Expect = 4e-14
Identities = 45/197 (22%), Positives = 93/197 (46%), Gaps = 1/197 (0%)
Frame = +2
Query: 333 EMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYANCL 392
E+ G+K D + C + K ++ K +H + ++ D + ++ +Y NC
Sbjct: 251 ELMEKGIKADANCFEILFDLCGKSKSVEDAKKVHDYFLQSTFRSDFKMHNKVIEMYGNCK 430
Query: 393 CVREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSILH 452
+ +A+ VFD MP+RN+ +W+ + Y N +GL +F +M G++ TML++L
Sbjct: 431 SMTDARRVFDHMPNRNMDSWHMMIRGYANSTMGDEGLQLFEQMNELGLEITSETMLAVLS 610
Query: 453 ACSDLQDLKSGKV-IHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHREVA 511
AC + ++ + + ++G+ V LL + + ++EA+ + +P
Sbjct: 611 ACGSAEAVEDAYIYLESMKSKYGIEPGVEHYMGLLDVLGQSGYLKEAEEFIEQLP----- 775
Query: 512 SWNGILTAYFTNKEYEK 528
+ +T + T K Y +
Sbjct: 776 -FEPTVTVFETLKNYAR 823
Score = 72.4 bits (176), Expect = 8e-13
Identities = 40/181 (22%), Positives = 85/181 (46%), Gaps = 1/181 (0%)
Frame = +2
Query: 232 EMVLDGVKPDPVTVSCILSACSDLQDLKSGKAIHGFALKHGMVENVFVSNALVNLYESCL 291
E++ G+K D + C + ++ K +H + L+ + + N ++ +Y +C
Sbjct: 251 ELMEKGIKADANCFEILFDLCGKSKSVEDAKKVHDYFLQSTFRSDFKMHNKVIEMYGNCK 430
Query: 292 CVREAQAVFDLMPHRNVITWNSLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILP 351
+ +A+ VFD MP+RN+ +W+ + Y N +GL +F +M G++ M ++L
Sbjct: 431 SMTDARRVFDHMPNRNMDSWHMMIRGYANSTMGDEGLQLFEQMNELGLEITSETMLAVLS 610
Query: 352 ACSQLKDLKSGKT-IHGFAVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMPHRNVV 410
AC + ++ + K+G+ V L+++ ++EA+ + +P V
Sbjct: 611 ACGSAEAVEDAYIYLESMKSKYGIEPGVEHYMGLLDVLGQSGYLKEAEEFIEQLPFEPTV 790
Query: 411 T 411
T
Sbjct: 791 T 793
Score = 68.2 bits (165), Expect = 1e-11
Identities = 42/177 (23%), Positives = 78/177 (43%), Gaps = 1/177 (0%)
Frame = +2
Query: 34 RGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSIGNAFIHAYGKCKCVEG 93
+GIK D F + C S+ K+ HD + SD + N I YG CK +
Sbjct: 263 KGIKADANCFEILFDLCGKSKSVEDAKKVHDYFLQSTFRSDFKMHNKVIEMYGNCKSMTD 442
Query: 94 ARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVKANPLTVSSILPGCSD 153
ARRVFD + R++ +W+ + Y N +GL +F +M ++ T+ ++L C
Sbjct: 443 ARRVFDHMPNRNMDSWHMMIRGYANSTMGDEGLQLFEQMNELGLEITSETMLAVLSACGS 622
Query: 154 LQDLKSGK-EIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVT 209
+ ++ + ++G+ V ++ + ++EA+ + +P VT
Sbjct: 623 AEAVEDAYIYLESMKSKYGIEPGVEHYMGLLDVLGQSGYLKEAEEFIEQLPFEPTVT 793
Score = 65.9 bits (159), Expect = 7e-11
Identities = 38/169 (22%), Positives = 77/169 (45%), Gaps = 1/169 (0%)
Frame = +2
Query: 434 EMVLNGVKPDLVTMLSILHACSDLQDLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCV 493
E++ G+K D + C + ++ K +H + ++ D + N ++ +Y C
Sbjct: 251 ELMEKGIKADANCFEILFDLCGKSKSVEDAKKVHDYFLQSTFRSDFKMHNKVIEMYGNCK 430
Query: 494 CVREAQVVFDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIG 553
+ +A+ VFD +P+R + SW+ ++ Y + ++GL +F QMN ++ T V+
Sbjct: 431 SMTDARRVFDHMPNRNMDSWHMMIRGYANSTMGDEGLQLFEQMNELGLEITSETMLAVLS 610
Query: 554 GCVKNSRIEEAMEIFRKMQT-MGFKPDETTIYSILRACSLSECLRMGKE 601
C +E+A M++ G +P +L S L+ +E
Sbjct: 611 ACGSAEAVEDAYIYLESMKSKYGIEPGVEHYMGLLDVLGQSGYLKEAEE 757
Score = 62.8 bits (151), Expect = 6e-10
Identities = 36/175 (20%), Positives = 81/175 (45%), Gaps = 1/175 (0%)
Frame = +2
Query: 137 VKANPLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQ 196
+KA+ + C + ++ K++H + ++ D + + + Y C + +A+
Sbjct: 269 IKADANCFEILFDLCGKSKSVEDAKKVHDYFLQSTFRSDFKMHNKVIEMYGNCKSMTDAR 448
Query: 197 TVFDLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQ 256
VFD MP+R++ +W+ + Y N +GL +F +M G++ T+ +LSAC +
Sbjct: 449 RVFDHMPNRNMDSWHMMIRGYANSTMGDEGLQLFEQMNELGLEITSETMLAVLSACGSAE 628
Query: 257 DLKSGKA-IHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVIT 310
++ + K+G+ V L+++ ++EA+ + +P +T
Sbjct: 629 AVEDAYIYLESMKSKYGIEPGVEHYMGLLDVLGQSGYLKEAEEFIEQLPFEPTVT 793
>BG455368 weakly similar to GP|9759524|dbj selenium-binding protein-like
{Arabidopsis thaliana}, partial (33%)
Length = 687
Score = 202 bits (514), Expect = 5e-52
Identities = 95/221 (42%), Positives = 140/221 (62%), Gaps = 1/221 (0%)
Frame = +1
Query: 733 LEEAYGFIQRMPMEPTAIAWKAFLAGCRVYKNVELAKISAKKLFEIDPNGSANYVTLFNI 792
+E+A +Q MPMEP W A L ++ N ++A+I+++ LFE++P+ NY+ L
Sbjct: 7 VEKALQLVQTMPMEPNGGVWGALLGASHIHGNPDVAEIASRSLFELEPDNLGNYLLLSKT 186
Query: 793 LVTAKLWSEASKIRKLMKERGITKTPGCSWFHVGNRV-HTFVAGDKSNMESDKIYNFLDE 851
A W + S++RKLM+E+ + K PGCSW N + H F AGD + E ++I LD+
Sbjct: 187 YALAAKWDDVSRVRKLMREKQLRKNPGCSWVEAKNGIIHEFFAGDVKHPEINEIKKALDD 366
Query: 852 LFAKIKAAGYKPDTDYVLHDIDQEEKAESLCNHSEKLAVAFGILNLNGQSTIRVFKNLRI 911
L ++K GY+P + V +DID E K L +HSEKLA+A+G+L+ + STI++ KNLRI
Sbjct: 367 LLQRLKCTGYQPKLNSVPYDIDDEGKRCLLVSHSEKLALAYGLLSTDAGSTIKIMKNLRI 546
Query: 912 CGDCHNAIKYMSNVVGVTIVVRDSLRFHHFKNGNCSCKDFW 952
C DCH + S + G I+VRD++ FHHF NG CSC +FW
Sbjct: 547 CEDCHIVMCGASXLTGRKIIVRDNMXFHHFLNGACSCNNFW 669
>BF633384 similar to PIR|T46179|T46 hypothetical protein T8H10.30 -
Arabidopsis thaliana, partial (22%)
Length = 595
Score = 162 bits (409), Expect = 7e-40
Identities = 77/197 (39%), Positives = 123/197 (62%), Gaps = 6/197 (3%)
Frame = +1
Query: 648 SWNTMIFANGMHGNGKEALSLFEKMLLS-----MVKPDSATFTCVLSACSHSMLVEEGVQ 702
+WN +I A GMHG G+EAL LF +M+ ++P+ T+ + ++ SHS +V+EG+
Sbjct: 4 TWNVLIMAYGMHGKGEEALKLFRRMVEEGDNNREIRPNEVTYIAIFASLSHSGMVDEGLN 183
Query: 703 IFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAYGFIQRMPMEPTAI-AWKAFLAGCRV 761
+F +M H +EP ++HY C+VD+ R+G +EEAY I+ MP + AW + L C++
Sbjct: 184 LFYTMKAKHGIEPTSDHYACLVDLLGRSGQIEEAYNLIKTMPSNMKKVDAWSSLLGACKI 363
Query: 762 YKNVELAKISAKKLFEIDPNGSANYVTLFNILVTAKLWSEASKIRKLMKERGITKTPGCS 821
++N+E+ +I+AK LF +DPN ++ YV L NI +A LW +A +RK MKE+G+ K
Sbjct: 364 HQNLEIGEIAAKNLFVLDPNVASYYVLLSNIYSSAGLWDQAIDVRKKMKEKGVRKNLDVV 543
Query: 822 WFHVGNRVHTFVAGDKS 838
++ R +F GD S
Sbjct: 544 GLNMAMRFTSFWQGDVS 594
Score = 44.3 bits (103), Expect = 2e-04
Identities = 40/152 (26%), Positives = 76/152 (49%), Gaps = 8/152 (5%)
Frame = +1
Query: 546 ITWSVVIGGCVKNSRIEEAMEIFRKMQTMG-----FKPDETTIYSILRACSLSECLRMGK 600
ITW+V+I + + EEA+++FR+M G +P+E T +I + S S + G
Sbjct: 1 ITWNVLIMAYGMHGKGEEALKLFRRMVEEGDNNREIRPNEVTYIAIFASLSHSGMVDEGL 180
Query: 601 EI-HCYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMP--IKDVFSWNTMIFANG 657
+ + +H + LVD+ + G + + N+ MP +K V +W++++ A
Sbjct: 181 NLFYTMKAKHGIEPTSDHYACLVDLLGRSGQIEEAYNLIKTMPSNMKKVDAWSSLLGACK 360
Query: 658 MHGNGKEALSLFEKMLLSMVKPDSATFTCVLS 689
+H N E + K L ++ P+ A++ +LS
Sbjct: 361 IHQN-LEIGEIAAKNLF-VLDPNVASYYVLLS 450
Score = 30.8 bits (68), Expect = 2.6
Identities = 26/115 (22%), Positives = 48/115 (41%), Gaps = 8/115 (6%)
Frame = +1
Query: 107 VTWNSLSACYVNCGFPQQGLNVFRKM-----GLNKVKANPLTVSSILPGCSDLQDLKSGK 161
+TWN L Y G ++ L +FR+M +++ N +T +I S + G
Sbjct: 1 ITWNVLIMAYGMHGKGEEALKLFRRMVEEGDNNREIRPNEVTYIAIFASLSHSGMVDEGL 180
Query: 162 EI-HGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPH--RDVVTWNSL 213
+ + +HG+ + V+ + + EA + MP + V W+SL
Sbjct: 181 NLFYTMKAKHGIEPTSDHYACLVDLLGRSGQIEEAYNLIKTMPSNMKKVDAWSSL 345
>AW776322 weakly similar to PIR|H84859|H84 hypothetical protein At2g42920
[imported] - Arabidopsis thaliana, partial (27%)
Length = 633
Score = 158 bits (399), Expect = 1e-38
Identities = 75/207 (36%), Positives = 128/207 (61%), Gaps = 1/207 (0%)
Frame = +1
Query: 642 PIKDVFSWNTMIFANGMHGNGKEALSLFEKMLLS-MVKPDSATFTCVLSACSHSMLVEEG 700
P + + WN++I M+G+ +EA F K+ S ++KPDS +F VL+AC H + +
Sbjct: 10 PRRGLSCWNSIIIGLAMNGHEREAFEFFSKLESSKLLKPDSVSFIGVLTACKHLGAINKA 189
Query: 701 VQIFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAYGFIQRMPMEPTAIAWKAFLAGCR 760
F M + +EP +HYTC+VD+ +AG LEEA I+ MP++P AI W + L+ CR
Sbjct: 190 RDYFELMMNKYEIEPSIKHYTCIVDVLGQAGLLEEAEELIKGMPLKPDAIIWGSLLSSCR 369
Query: 761 VYKNVELAKISAKKLFEIDPNGSANYVTLFNILVTAKLWSEASKIRKLMKERGITKTPGC 820
++NV++A+ +A++++E++P+ ++ YV + N+ + + EA + R LMKE K PGC
Sbjct: 370 KHRNVQIARRAAQRVYELNPSDASGYVLMSNVHAASNKFEEAIEQRLLMKENLTEKEPGC 549
Query: 821 SWFHVGNRVHTFVAGDKSNMESDKIYN 847
S + VH F+AG + + ++ +IY+
Sbjct: 550 SSIELYGEVHEFIAGGRLHPKTQEIYH 630
Score = 35.4 bits (80), Expect = 0.10
Identities = 34/157 (21%), Positives = 65/157 (40%), Gaps = 4/157 (2%)
Frame = +1
Query: 304 PHRNVITWNSLASCYVNCGFPQKGLNVFREMGLNGV-KPDPMAMSSILPACSQLKDLKSG 362
P R + WNS+ G ++ F ++ + + KPD ++ +L AC L +
Sbjct: 10 PRRGLSCWNSIIIGLAMNGHEREAFEFFSKLESSKLLKPDSVSFIGVLTACKHLGAINKA 189
Query: 363 KTIHGFAV-KHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMPHR-NVVTWNS-LSSCY 419
+ + K+ + + T +V++ + EA+ + MP + + + W S LSSC
Sbjct: 190 RDYFELMMNKYEIEPSIKHYTCIVDVLGQAGLLEEAEELIKGMPLKPDAIIWGSLLSSCR 369
Query: 420 VNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHACSD 456
+ R LN ++S +HA S+
Sbjct: 370 KHRNVQIARRAAQRVYELNPSDASGYVLMSNVHAASN 480
Score = 31.2 bits (69), Expect = 2.0
Identities = 26/117 (22%), Positives = 51/117 (43%), Gaps = 3/117 (2%)
Frame = +1
Query: 203 PHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGV-KPDPVTVSCILSACSDLQDLKSG 261
P R + WNS+ G ++ F ++ + KPD V+ +L+AC L +
Sbjct: 10 PRRGLSCWNSIIIGLAMNGHEREAFEFFSKLESSKLLKPDSVSFIGVLTACKHLGAINKA 189
Query: 262 KAIHGFAL-KHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHR-NVITWNSLAS 316
+ + K+ + ++ +V++ + EA+ + MP + + I W SL S
Sbjct: 190 RDYFELMMNKYEIEPSIKHYTCIVDVLGQAGLLEEAEELIKGMPLKPDAIIWGSLLS 360
>CA921534 similar to PIR|D96516|D96 F16N3.14 [imported] - Arabidopsis
thaliana, partial (18%)
Length = 790
Score = 157 bits (397), Expect = 2e-38
Identities = 89/235 (37%), Positives = 133/235 (55%)
Frame = -1
Query: 718 EHYTCVVDIYSRAGCLEEAYGFIQRMPMEPTAIAWKAFLAGCRVYKNVELAKISAKKLFE 777
E Y VV+I+ A L +A+ F++ MP+EP W+ R++ ++E + + L
Sbjct: 784 EPYLGVVNIFGCAVGLMKAHEFMRNMPIEPGVELWETLRNFARIHGDLEREDCADELLTV 605
Query: 778 IDPNGSANYVTLFNILVTAKLWSEASKIRKLMKERGITKTPGCSWFHVGNRVHTFVAGDK 837
+DP+ +A A K+ ++ K + NRV +
Sbjct: 604 LDPSKAA-----------------ADKVPLPQRK----KQSAINMLEEKNRVSEYRCNMP 488
Query: 838 SNMESDKIYNFLDELFAKIKAAGYKPDTDYVLHDIDQEEKAESLCNHSEKLAVAFGILNL 897
E D L L +++ AGY PDT YVLHDID+EEK ++L HSE+LA+A+G+++
Sbjct: 487 YKEEGDVK---LRGLTGQMREAGYVPDTRYVLHDIDEEEKEKALQYHSERLAIAYGLIST 317
Query: 898 NGQSTIRVFKNLRICGDCHNAIKYMSNVVGVTIVVRDSLRFHHFKNGNCSCKDFW 952
++T+R+ KNLRICGDCHNAIK MS +VG ++VRD+ RFHHFK+G CSC D+W
Sbjct: 316 PPRTTLRIIKNLRICGDCHNAIKIMSKIVGRELIVRDNKRFHHFKDGKCSCGDYW 152
>AL370459 weakly similar to GP|20146256|d hypothetical protein {Oryza sativa
(japonica cultivar-group)}, partial (11%)
Length = 477
Score = 156 bits (394), Expect = 4e-38
Identities = 72/126 (57%), Positives = 91/126 (72%)
Frame = +1
Query: 827 NRVHTFVAGDKSNMESDKIYNFLDELFAKIKAAGYKPDTDYVLHDIDQEEKAESLCNHSE 886
N+VH F GD + ++ +IY LDEL KIK GY P+TD+VLHD++ E+K + L HSE
Sbjct: 4 NQVHKFHVGDTLHPKAQQIYEKLDELALKIKNVGYVPNTDFVLHDVEDEQKEQYLFQHSE 183
Query: 887 KLAVAFGILNLNGQSTIRVFKNLRICGDCHNAIKYMSNVVGVTIVVRDSLRFHHFKNGNC 946
KLAVAF +++ IRVFKNLR+CGDCH AIKY+S V G IVVRD+ RFHH K+G C
Sbjct: 184 KLAVAFALISTPNPKPIRVFKNLRVCGDCHTAIKYISMVSGREIVVRDANRFHHMKDGTC 363
Query: 947 SCKDFW 952
SC D+W
Sbjct: 364 SCNDYW 381
>BE941714 weakly similar to GP|11994734|dbj selenium-binding protein-like
{Arabidopsis thaliana}, partial (10%)
Length = 619
Score = 150 bits (378), Expect = 3e-36
Identities = 77/198 (38%), Positives = 122/198 (60%), Gaps = 5/198 (2%)
Frame = +2
Query: 618 TNALVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGKEALSLFEKMLLSMV 677
+ +L+DMYAKCG L L++ +FD M ++DV WN MI MHG+GK AL LF M V
Sbjct: 23 STSLLDMYAKCGNLELAKRLFDSMNMRDVVCWNAMISGMAMHGDGKGALKLFYDMEKVGV 202
Query: 678 KPDSATFTCVLSACSHSMLVEEGVQIFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAY 737
KPD TF V +ACS+S + EG+ + + M + + P++EHY C+VD+ SRAG EEA
Sbjct: 203 KPDDITFIAVFTACSYSGMAYEGLMLLDKMCSVYNIVPKSEHYGCLVDLLSRAGLFEEAM 382
Query: 738 GFIQRMPM----EPTAIAWKAFLAGCRVYKNVELAKISAKKLFEIDPN-GSANYVTLFNI 792
I+++ +AW+AFL+ C + +LA+++A+K+ ++D + S YV L N+
Sbjct: 383 VMIRKITNSWNGSEETLAWRAFLSACCNHGETQLAELAAEKVLQLDNHIHSGVYVLLSNL 562
Query: 793 LVTAKLWSEASKIRKLMK 810
+ ++A +++ +MK
Sbjct: 563 YAASGKHNDARRVKDMMK 616
Score = 58.2 bits (139), Expect = 2e-08
Identities = 48/161 (29%), Positives = 72/161 (43%), Gaps = 11/161 (6%)
Frame = +2
Query: 176 VFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVL 235
V +S++ ++ YAKC + A+ +FD M RDVV WN++ S G + L +F +M
Sbjct: 14 VRLSTSLLDMYAKCGNLELAKRLFDSMNMRDVVCWNAMISGMAMHGDGKGALKLFYDMEK 193
Query: 236 DGVKPDPVTVSCILSACSDLQDLKSGKAIHGFALKHGM--VENVFVSN----ALVNLYES 289
GVKPD +T + +ACS SG A G L M V N+ + LV+L
Sbjct: 194 VGVKPDDITFIAVFTACS-----YSGMAYEGLMLLDKMCSVYNIVPKSEHYGCLVDLLSR 358
Query: 290 CLCVREAQAVFDLMPH-----RNVITWNSLASCYVNCGFPQ 325
EA + + + + W + S N G Q
Sbjct: 359 AGLFEEAMVMIRKITNSWNGSEETLAWRAFLSACCNHGETQ 481
Score = 56.6 bits (135), Expect = 4e-08
Identities = 43/144 (29%), Positives = 68/144 (46%), Gaps = 6/144 (4%)
Frame = +2
Query: 378 VFVCTALVNLYANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVL 437
V + T+L+++YA C + A+ +FD M R+VV WN++ S G + L +F +M
Sbjct: 14 VRLSTSLLDMYAKCGNLELAKRLFDSMNMRDVVCWNAMISGMAMHGDGKGALKLFYDMEK 193
Query: 438 NGVKPDLVTMLSILHACSDLQDLKSGKVIHGFAVRHGMVEDVFVC------NALLSLYAK 491
GVKPD +T +++ ACS SG G + M + L+ L ++
Sbjct: 194 VGVKPDDITFIAVFTACS-----YSGMAYEGLMLLDKMCSVYNIVPKSEHYGCLVDLLSR 358
Query: 492 CVCVREAQVVFDLIPHREVASWNG 515
EA V+ I + SWNG
Sbjct: 359 AGLFEEAMVMIRKITN----SWNG 418
Score = 50.4 bits (119), Expect = 3e-06
Identities = 26/79 (32%), Positives = 45/79 (56%)
Frame = +2
Query: 276 NVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASCYVNCGFPQKGLNVFREMG 335
+V +S +L+++Y C + A+ +FD M R+V+ WN++ S G + L +F +M
Sbjct: 11 SVRLSTSLLDMYAKCGNLELAKRLFDSMNMRDVVCWNAMISGMAMHGDGKGALKLFYDME 190
Query: 336 LNGVKPDPMAMSSILPACS 354
GVKPD + ++ ACS
Sbjct: 191 KVGVKPDDITFIAVFTACS 247
Score = 49.3 bits (116), Expect = 7e-06
Identities = 27/93 (29%), Positives = 48/93 (51%)
Frame = +2
Query: 479 VFVCNALLSLYAKCVCVREAQVVFDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQMNR 538
V + +LL +YAKC + A+ +FD + R+V WN +++ + + + L +F M +
Sbjct: 14 VRLSTSLLDMYAKCGNLELAKRLFDSMNMRDVVCWNAMISGMAMHGDGKGALKLFYDMEK 193
Query: 539 DEVKADEITWSVVIGGCVKNSRIEEAMEIFRKM 571
VK D+IT+ V C + E + + KM
Sbjct: 194 VGVKPDDITFIAVFTACSYSGMAYEGLMLLDKM 292
Score = 45.4 bits (106), Expect = 1e-04
Identities = 24/78 (30%), Positives = 40/78 (50%)
Frame = +2
Query: 75 VSIGNAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGL 134
V + + + Y KC +E A+R+FD + RDVV WN++ + G + L +F M
Sbjct: 14 VRLSTSLLDMYAKCGNLELAKRLFDSMNMRDVVCWNAMISGMAMHGDGKGALKLFYDMEK 193
Query: 135 NKVKANPLTVSSILPGCS 152
VK + +T ++ CS
Sbjct: 194 VGVKPDDITFIAVFTACS 247
Score = 41.2 bits (95), Expect = 0.002
Identities = 22/78 (28%), Positives = 38/78 (48%)
Frame = +2
Query: 516 ILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGGCVKNSRIEEAMEIFRKMQTMG 575
+L Y E +F MN +V + W+ +I G + + A+++F M+ +G
Sbjct: 32 LLDMYAKCGNLELAKRLFDSMNMRDV----VCWNAMISGMAMHGDGKGALKLFYDMEKVG 199
Query: 576 FKPDETTIYSILRACSLS 593
KPD+ T ++ ACS S
Sbjct: 200 VKPDDITFIAVFTACSYS 253
Score = 30.0 bits (66), Expect = 4.4
Identities = 14/36 (38%), Positives = 20/36 (54%)
Frame = +2
Query: 18 HGLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAAS 53
HG A+K++ G+KPD F+AV AC+ S
Sbjct: 146 HGDGKGALKLFYDMEKVGVKPDDITFIAVFTACSYS 253
>TC83482 weakly similar to GP|9758453|dbj|BAB08982.1 selenium-binding
protein-like {Arabidopsis thaliana}, partial (10%)
Length = 895
Score = 139 bits (351), Expect = 4e-33
Identities = 70/178 (39%), Positives = 103/178 (57%)
Frame = +2
Query: 645 DVFSWNTMIFANGMHGNGKEALSLFEKMLLSMVKPDSATFTCVLSACSHSMLVEEGVQIF 704
D+ SWN +I HG AL F++M + + PD TF VLSAC H+ LVEEG + F
Sbjct: 2 DLVSWNAIIGGYASHGLATRALEEFDRMKV-VGTPDEVTFVNVLSACVHAGLVEEGEKHF 178
Query: 705 NSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAYGFIQRMPMEPTAIAWKAFLAGCRVYKN 764
M + ++ E EHY+C+VD+Y RAG +EA I+ MP EP + W A LA C ++ N
Sbjct: 179 TDMLTKYGIQAEMEHYSCMVDLYGRAGRFDEAENLIKNMPFEPDVVLWGALLAACGLHSN 358
Query: 765 VELAKISAKKLFEIDPNGSANYVTLFNILVTAKLWSEASKIRKLMKERGITKTPGCSW 822
+EL + +A+++ ++ + +Y L I +WS +++R MKERGI K SW
Sbjct: 359 LELGEYAAERIRRLESSHPVSYSVLSKIQGEKGVWSSVNELRDTMKERGIKKQTAISW 532
Score = 60.1 bits (144), Expect = 4e-09
Identities = 33/120 (27%), Positives = 61/120 (50%), Gaps = 2/120 (1%)
Frame = +2
Query: 544 DEITWSVVIGGCVKNSRIEEAMEIFRKMQTMGFKPDETTIYSILRACSLSECLRMG-KEI 602
D ++W+ +IGG + A+E F +M+ +G PDE T ++L AC + + G K
Sbjct: 2 DLVSWNAIIGGYASHGLATRALEEFDRMKVVG-TPDEVTFVNVLSACVHAGLVEEGEKHF 178
Query: 603 HCYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIK-DVFSWNTMIFANGMHGN 661
+ ++ ++ + +VD+Y + G + N+ MP + DV W ++ A G+H N
Sbjct: 179 TDMLTKYGIQAEMEHYSCMVDLYGRAGRFDEAENLIKNMPFEPDVVLWGALLAACGLHSN 358
>BI308000 similar to GP|7363286|dbj| Similar to Arabidopsis thaliana DNA
chromosome 4 BAC clone F4I10; vegetative storage
protein., partial (12%)
Length = 771
Score = 133 bits (335), Expect = 3e-31
Identities = 62/111 (55%), Positives = 83/111 (73%)
Frame = +3
Query: 842 SDKIYNFLDELFAKIKAAGYKPDTDYVLHDIDQEEKAESLCNHSEKLAVAFGILNLNGQS 901
S++IY FL+ L KI+ AGY PDT+ + HD++++ K + L +HSE+LA+AFG+LN N +
Sbjct: 6 SEEIYAFLEALGDKIRDAGYIPDTNSI-HDVEEKVKEQLLSSHSERLAIAFGLLNTNHGT 182
Query: 902 TIRVFKNLRICGDCHNAIKYMSNVVGVTIVVRDSLRFHHFKNGNCSCKDFW 952
I V KNLR+CGDCH+ KY+S V G I+VRD RFHHFKNG CSC D+W
Sbjct: 183 PIHVRKNLRVCGDCHDVTKYISLVTGREIIVRDLRRFHHFKNGICSCGDYW 335
>AW774626 similar to PIR|T47909|T47 hypothetical protein T20K12.70 -
Arabidopsis thaliana, partial (12%)
Length = 703
Score = 131 bits (329), Expect = 1e-30
Identities = 77/234 (32%), Positives = 122/234 (51%), Gaps = 2/234 (0%)
Frame = +3
Query: 82 IHAYGKCKCVEGARRVFDDLV--ARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVKA 139
+ Y KCKCV A +F L ++ V W ++ Y G + + FR M V+
Sbjct: 6 VDMYAKCKCVSEAEFLFKGLEFDRKNHVLWTAMVTGYAQNGDGYKAVEFFRYMHAQGVEC 185
Query: 140 NPLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVF 199
N T +IL CS + G+++HGF+V+ G +V+V SA V+ YAKC ++ A+ +
Sbjct: 186 NQYTFPTILTACSSVLARCFGEQVHGFIVKSGFGSNVYVQSALVDMYAKCGDLKNAKNML 365
Query: 200 DLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQDLK 259
+ M DVV+WNSL +V G ++ L +F+ M +K D T +L+ C +
Sbjct: 366 ETMEDDDVVSWNSLMVGFVRHGLEEEALRLFKNMHGRNMKIDDYTFPSVLNCC--VVGSI 539
Query: 260 SGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNS 313
+ K++HG +K G VSNALV++Y + A VF+ M ++VI+W S
Sbjct: 540 NPKSVHGLIIKTGFENYKLVSNALVDMYAKTGDMDCAYTVFEKMLEKDVISWTS 701
Score = 130 bits (326), Expect = 3e-30
Identities = 76/234 (32%), Positives = 125/234 (52%), Gaps = 2/234 (0%)
Frame = +3
Query: 183 VNFYAKCLCVREAQTVFDLMP--HRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKP 240
V+ YAKC CV EA+ +F + ++ V W ++ + Y G K + FR M GV+
Sbjct: 6 VDMYAKCKCVSEAEFLFKGLEFDRKNHVLWTAMVTGYAQNGDGYKAVEFFRYMHAQGVEC 185
Query: 241 DPVTVSCILSACSDLQDLKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVF 300
+ T IL+ACS + G+ +HGF +K G NV+V +ALV++Y C ++ A+ +
Sbjct: 186 NQYTFPTILTACSSVLARCFGEQVHGFIVKSGFGSNVYVQSALVDMYAKCGDLKNAKNML 365
Query: 301 DLMPHRNVITWNSLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLK 360
+ M +V++WNSL +V G ++ L +F+ M +K D S+L C +
Sbjct: 366 ETMEDDDVVSWNSLMVGFVRHGLEEEALRLFKNMHGRNMKIDDYTFPSVLNCC--VVGSI 539
Query: 361 SGKTIHGFAVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMPHRNVVTWNS 414
+ K++HG +K G V ALV++YA + A TVF+ M ++V++W S
Sbjct: 540 NPKSVHGLIIKTGFENYKLVSNALVDMYAKTGDMDCAYTVFEKMLEKDVISWTS 701
Score = 127 bits (319), Expect = 2e-29
Identities = 75/233 (32%), Positives = 124/233 (53%), Gaps = 2/233 (0%)
Frame = +3
Query: 283 LVNLYESCLCVREAQAVFDLMP--HRNVITWNSLASCYVNCGFPQKGLNVFREMGLNGVK 340
LV++Y C CV EA+ +F + +N + W ++ + Y G K + FR M GV+
Sbjct: 3 LVDMYAKCKCVSEAEFLFKGLEFDRKNHVLWTAMVTGYAQNGDGYKAVEFFRYMHAQGVE 182
Query: 341 PDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYANCLCVREAQTV 400
+ +IL ACS + G+ +HGF VK G +V+V +ALV++YA C ++ A+ +
Sbjct: 183 CNQYTFPTILTACSSVLARCFGEQVHGFIVKSGFGSNVYVQSALVDMYAKCGDLKNAKNM 362
Query: 401 FDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHACSDLQDL 460
+ M +VV+WNSL +V G ++ L +F+ M +K D T S+L+ C +
Sbjct: 363 LETMEDDDVVSWNSLMVGFVRHGLEEEALRLFKNMHGRNMKIDDYTFPSVLNCC--VVGS 536
Query: 461 KSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHREVASW 513
+ K +HG ++ G V NAL+ +YAK + A VF+ + ++V SW
Sbjct: 537 INPKSVHGLIIKTGFENYKLVSNALVDMYAKTGDMDCAYTVFEKMLEKDVISW 695
Score = 117 bits (293), Expect = 2e-26
Identities = 78/270 (28%), Positives = 127/270 (46%), Gaps = 2/270 (0%)
Frame = +3
Query: 384 LVNLYANCLCVREAQTVFDLMP--HRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVK 441
LV++YA C CV EA+ +F + +N V W ++ + Y G K + FR M GV+
Sbjct: 3 LVDMYAKCKCVSEAEFLFKGLEFDRKNHVLWTAMVTGYAQNGDGYKAVEFFRYMHAQGVE 182
Query: 442 PDLVTMLSILHACSDLQDLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVV 501
+ T +IL ACS + G+ +HGF V+ G +V+V +AL+ +YAKC ++ A+ +
Sbjct: 183 CNQYTFPTILTACSSVLARCFGEQVHGFIVKSGFGSNVYVQSALVDMYAKCGDLKNAKNM 362
Query: 502 FDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGGCVKNSRI 561
+ + +V SWN ++ + + E+ L +F M+ +K D+ T+ V+ CV S
Sbjct: 363 LETMEDDDVVSWNSLMVGFVRHGLEEEALRLFKNMHGRNMKIDDYTFPSVLNCCVVGS-- 536
Query: 562 EEAMEIFRKMQTMGFKPDETTIYSILRACSLSECLRMGKEIHCYVFRHWKDWDLARTNAL 621
P K +H + + + +NAL
Sbjct: 537 --------------INP---------------------KSVHGLIIKTGFENYKLVSNAL 611
Query: 622 VDMYAKCGGLSLSRNVFDMMPIKDVFSWNT 651
VDMYAK G + + VF+ M KDV SW +
Sbjct: 612 VDMYAKTGDMDCAYTVFEKMLEKDVISWTS 701
Score = 110 bits (276), Expect = 2e-24
Identities = 78/277 (28%), Positives = 131/277 (47%), Gaps = 9/277 (3%)
Frame = +3
Query: 485 LLSLYAKCVCVREAQVVFDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQMNRDEVKAD 544
L+ +YAKC CV EA+ +F KGL F + N
Sbjct: 3 LVDMYAKCKCVSEAEFLF-------------------------KGLE-FDRKNH------ 86
Query: 545 EITWSVVIGGCVKNSRIEEAMEIFRKMQTMGFKPDETTIYSILRACSLSECLRMGKEIHC 604
+ W+ ++ G +N +A+E FR M G + ++ T +IL ACS G+++H
Sbjct: 87 -VLWTAMVTGYAQNGDGYKAVEFFRYMHAQGVECNQYTFPTILTACSSVLARCFGEQVHG 263
Query: 605 YVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGKE 664
++ + ++ +ALVDMYAKCG L ++N+ + M DV SWN+++ HG +E
Sbjct: 264 FIVKSGFGSNVYVQSALVDMYAKCGDLKNAKNMLETMEDDDVVSWNSLMVGFVRHGLEEE 443
Query: 665 ALSLFEKMLLSMVKPDSATFTCVLSAC---------SHSMLVEEGVQIFNSMSRDHLVEP 715
AL LF+ M +K D TF VL+ C H ++++ G + + +S
Sbjct: 444 ALRLFKNMHGRNMKIDDYTFPSVLNCCVVGSINPKSVHGLIIKTGFENYKLVS------- 602
Query: 716 EAEHYTCVVDIYSRAGCLEEAYGFIQRMPMEPTAIAW 752
+VD+Y++ G ++ AY ++M +E I+W
Sbjct: 603 -----NALVDMYAKTGDMDCAYTVFEKM-LEKDVISW 695
Score = 94.4 bits (233), Expect = 2e-19
Identities = 53/190 (27%), Positives = 99/190 (51%)
Frame = +3
Query: 23 EAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSIGNAFI 82
+A++ + A+G++ ++ F + AC++ +Q H + G S+V + +A +
Sbjct: 138 KAVEFFRYMHAQGVECNQYTFPTILTACSSVLARCFGEQVHGFIVKSGFGSNVYVQSALV 317
Query: 83 HAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVKANPL 142
Y KC ++ A+ + + + DVV+WNSL +V G ++ L +F+ M +K +
Sbjct: 318 DMYAKCGDLKNAKNMLETMEDDDVVSWNSLMVGFVRHGLEEEALRLFKNMHGRNMKIDDY 497
Query: 143 TVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLM 202
T S+L C + + K +HG +++ G VS+A V+ YAK + A TVF+ M
Sbjct: 498 TFPSVLNCC--VVGSINPKSVHGLIIKTGFENYKLVSNALVDMYAKTGDMDCAYTVFEKM 671
Query: 203 PHRDVVTWNS 212
+DV++W S
Sbjct: 672 LEKDVISWTS 701
Score = 53.5 bits (127), Expect = 4e-07
Identities = 36/140 (25%), Positives = 67/140 (47%), Gaps = 2/140 (1%)
Frame = +3
Query: 621 LVDMYAKCGGLSLSRNVFDMMPI--KDVFSWNTMIFANGMHGNGKEALSLFEKMLLSMVK 678
LVDMYAKC +S + +F + K+ W M+ +G+G +A+ F M V+
Sbjct: 3 LVDMYAKCKCVSEAEFLFKGLEFDRKNHVLWTAMVTGYAQNGDGYKAVEFFRYMHAQGVE 182
Query: 679 PDSATFTCVLSACSHSMLVEEGVQIFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAYG 738
+ TF +L+ACS + G Q+ + + + +VD+Y++ G L+ A
Sbjct: 183 CNQYTFPTILTACSSVLARCFGEQVHGFIVKSGF-GSNVYVQSALVDMYAKCGDLKNAKN 359
Query: 739 FIQRMPMEPTAIAWKAFLAG 758
++ M + ++W + + G
Sbjct: 360 MLETME-DDDVVSWNSLMVG 416
>BE202961 weakly similar to GP|6016735|gb|A hypothetical protein {Arabidopsis
thaliana}, partial (14%)
Length = 613
Score = 130 bits (326), Expect = 3e-30
Identities = 65/202 (32%), Positives = 113/202 (55%)
Frame = +2
Query: 126 LNVFRKMGLNKVKANPLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNF 185
L++ +M ++K N +TV S+L C+ + +L+ G +IH V +G + VS+A ++
Sbjct: 2 LDLSSEMLDKRIKPNWVTVVSVLRACACISNLEEGMKIHELAVNYGFEMETTVSTALMDM 181
Query: 186 YAKCLCVREAQTVFDLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTV 245
Y KC +A +F+ MP +DV+ W L S Y + G + + VFR M+ G +PD + +
Sbjct: 182 YMKCFSPEKAVDIFNRMPKKDVIAWAVLFSGYADNGMVHESMWVFRNMLSSGTRPDAIAL 361
Query: 246 SCILSACSDLQDLKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPH 305
IL+ S+L L+ H F +K+G N F+ +L+ +Y C + +A VF M +
Sbjct: 362 VKILTTVSELGILQQAVCFHAFVIKNGFENNQFIGASLIEVYAKCSSIEDANKVFKGMTY 541
Query: 306 RNVITWNSLASCYVNCGFPQKG 327
++V+TW+S+ + Y GF +G
Sbjct: 542 KDVVTWSSIIAAY---GFHGQG 598
Score = 130 bits (326), Expect = 3e-30
Identities = 69/202 (34%), Positives = 114/202 (56%)
Frame = +2
Query: 227 LNVFREMVLDGVKPDPVTVSCILSACSDLQDLKSGKAIHGFALKHGMVENVFVSNALVNL 286
L++ EM+ +KP+ VTV +L AC+ + +L+ G IH A+ +G VS AL+++
Sbjct: 2 LDLSSEMLDKRIKPNWVTVVSVLRACACISNLEEGMKIHELAVNYGFEMETTVSTALMDM 181
Query: 287 YESCLCVREAQAVFDLMPHRNVITWNSLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAM 346
Y C +A +F+ MP ++VI W L S Y + G + + VFR M +G +PD +A+
Sbjct: 182 YMKCFSPEKAVDIFNRMPKKDVIAWAVLFSGYADNGMVHESMWVFRNMLSSGTRPDAIAL 361
Query: 347 SSILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMPH 406
IL S+L L+ H F +K+G + F+ +L+ +YA C + +A VF M +
Sbjct: 362 VKILTTVSELGILQQAVCFHAFVIKNGFENNQFIGASLIEVYAKCSSIEDANKVFKGMTY 541
Query: 407 RNVVTWNSLSSCYVNCGFPQKG 428
++VVTW+S+ + Y GF +G
Sbjct: 542 KDVVTWSSIIAAY---GFHGQG 598
Score = 127 bits (320), Expect = 2e-29
Identities = 63/203 (31%), Positives = 116/203 (57%)
Frame = +2
Query: 328 LNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNL 387
L++ EM +KP+ + + S+L AC+ + +L+ G IH AV +G + V TAL+++
Sbjct: 2 LDLSSEMLDKRIKPNWVTVVSVLRACACISNLEEGMKIHELAVNYGFEMETTVSTALMDM 181
Query: 388 YANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTM 447
Y C +A +F+ MP ++V+ W L S Y + G + + VFR M+ +G +PD + +
Sbjct: 182 YMKCFSPEKAVDIFNRMPKKDVIAWAVLFSGYADNGMVHESMWVFRNMLSSGTRPDAIAL 361
Query: 448 LSILHACSDLQDLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPH 507
+ IL S+L L+ H F +++G + F+ +L+ +YAKC + +A VF + +
Sbjct: 362 VKILTTVSELGILQQAVCFHAFVIKNGFENNQFIGASLIEVYAKCSSIEDANKVFKGMTY 541
Query: 508 REVASWNGILTAYFTNKEYEKGL 530
++V +W+ I+ AY + + E+ L
Sbjct: 542 KDVVTWSSIIAAYGFHGQGEEAL 610
Score = 116 bits (291), Expect = 4e-26
Identities = 69/238 (28%), Positives = 114/238 (46%)
Frame = +2
Query: 429 LNVFREMVLNGVKPDLVTMLSILHACSDLQDLKSGKVIHGFAVRHGMVEDVFVCNALLSL 488
L++ EM+ +KP+ VT++S+L AC+ + +L+ G IH AV +G + V AL+ +
Sbjct: 2 LDLSSEMLDKRIKPNWVTVVSVLRACACISNLEEGMKIHELAVNYGFEMETTVSTALMDM 181
Query: 489 YAKCVCVREAQVVFDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQMNRDEVKADEITW 548
Y KC +A +F+ +P K D I W
Sbjct: 182 YMKCFSPEKAVDIFNRMP-----------------------------------KKDVIAW 256
Query: 549 SVVIGGCVKNSRIEEAMEIFRKMQTMGFKPDETTIYSILRACSLSECLRMGKEIHCYVFR 608
+V+ G N + E+M +FR M + G +PD + IL S L+ H +V +
Sbjct: 257 AVLFSGYADNGMVHESMWVFRNMLSSGTRPDAIALVKILTTVSELGILQQAVCFHAFVIK 436
Query: 609 HWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGKEAL 666
+ + + +L+++YAKC + + VF M KDV +W+++I A G HG G+EAL
Sbjct: 437 NGFENNQFIGASLIEVYAKCSSIEDANKVFKGMTYKDVVTWSSIIAAYGFHGQGEEAL 610
Score = 100 bits (248), Expect = 3e-21
Identities = 55/191 (28%), Positives = 99/191 (51%)
Frame = +2
Query: 36 IKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSIGNAFIHAYGKCKCVEGAR 95
IKP+ ++V +ACA + + + H+ A G + ++ A + Y KC E A
Sbjct: 35 IKPNWVTVVSVLRACACISNLEEGMKIHELAVNYGFEMETTVSTALMDMYMKCFSPEKAV 214
Query: 96 RVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVKANPLTVSSILPGCSDLQ 155
+F+ + +DV+ W L + Y + G + + VFR M + + + + + IL S+L
Sbjct: 215 DIFNRMPKKDVIAWAVLFSGYADNGMVHESMWVFRNMLSSGTRPDAIALVKILTTVSELG 394
Query: 156 DLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVTWNSLSS 215
L+ H FV+++G + F+ ++ + YAKC + +A VF M ++DVVTW+S+ +
Sbjct: 395 ILQQAVCFHAFVIKNGFENNQFIGASLIEVYAKCSSIEDANKVFKGMTYKDVVTWSSIIA 574
Query: 216 CYVNCGFPQKG 226
Y GF +G
Sbjct: 575 AY---GFHGQG 598
Score = 77.0 bits (188), Expect = 3e-14
Identities = 46/193 (23%), Positives = 94/193 (47%)
Frame = +2
Query: 565 MEIFRKMQTMGFKPDETTIYSILRACSLSECLRMGKEIHCYVFRHWKDWDLARTNALVDM 624
+++ +M KP+ T+ S+LRAC+ L G +IH + + + + AL+DM
Sbjct: 2 LDLSSEMLDKRIKPNWVTVVSVLRACACISNLEEGMKIHELAVNYGFEMETTVSTALMDM 181
Query: 625 YAKCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGKEALSLFEKMLLSMVKPDSATF 684
Y KC + ++F+ MP KDV +W + +G E++ +F ML S +PD+
Sbjct: 182 YMKCFSPEKAVDIFNRMPKKDVIAWAVLFSGYADNGMVHESMWVFRNMLSSGTRPDAIAL 361
Query: 685 TCVLSACSHSMLVEEGVQIFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAYGFIQRMP 744
+L+ S ++++ V F++ + E ++++Y++ +E+A + M
Sbjct: 362 VKILTTVSELGILQQAV-CFHAFVIKNGFENNQFIGASLIEVYAKCSSIEDANKVFKGMT 538
Query: 745 MEPTAIAWKAFLA 757
+ + W + +A
Sbjct: 539 YK-DVVTWSSIIA 574
>BF635667 weakly similar to PIR|C85359|C853 hypothetical protein AT4g30700
[imported] - Arabidopsis thaliana, partial (6%)
Length = 603
Score = 129 bits (324), Expect = 5e-30
Identities = 63/157 (40%), Positives = 98/157 (62%), Gaps = 1/157 (0%)
Frame = +3
Query: 684 FTCVLSACSHSMLVEEGVQIFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAYGFIQRM 743
F V++ C L+++G +IF SM D+ +EP AEHY C+VD++SR G LEEA ++RM
Sbjct: 6 F*LVVATCG---LLDQGKKIFGSMISDYGLEPTAEHYACMVDLFSRCGRLEEALQLLERM 176
Query: 744 PMEP-TAIAWKAFLAGCRVYKNVELAKISAKKLFEIDPNGSANYVTLFNILVTAKLWSEA 802
T W A LAGC +++NV++ +++A LF+++PN ++NYV L+ I + +
Sbjct: 177 KSSSVTGSMWGALLAGCVMHQNVKIGEVAAHHLFQLEPNNTSNYVALWGIYQSRGMVLGV 356
Query: 803 SKIRKLMKERGITKTPGCSWFHVGNRVHTFVAGDKSN 839
S I M++ G+ KTPGC W ++ R H F GD S+
Sbjct: 357 STIXGKMRDLGLVKTPGCXWINIAGRAHKFYQGDLSH 467
>BG645370 weakly similar to PIR|T00405|T004 hypothetical protein At2g44880
[imported] - Arabidopsis thaliana, partial (11%)
Length = 784
Score = 122 bits (307), Expect(2) = 1e-29
Identities = 54/138 (39%), Positives = 93/138 (67%)
Frame = -2
Query: 684 FTCVLSACSHSMLVEEGVQIFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAYGFIQRM 743
FT VL+AC+H+ +++G + FN M ++ + P+ + Y C++D+Y+R G L +A ++ M
Sbjct: 783 FTAVLTACNHAGFIDKGEEYFNKMITNYGLSPDIDIYACLIDLYARNGNLRKARDLMEEM 604
Query: 744 PMEPTAIAWKAFLAGCRVYKNVELAKISAKKLFEIDPNGSANYVTLFNILVTAKLWSEAS 803
P +P I W +FL+ C++Y +VEL + +A +L +++P +A Y+TL +I T LW+EAS
Sbjct: 603 PYDPNCIIWSSFLSACKIYGDVELGREAAIQLIKMEPCNAAPYLTLAHIYTTKGLWNEAS 424
Query: 804 KIRKLMKERGITKTPGCS 821
++R LM++R K PG S
Sbjct: 423 EVRSLMQQRVKRKPPGWS 370
Score = 34.3 bits (77), Expect = 0.23
Identities = 20/77 (25%), Positives = 43/77 (54%), Gaps = 3/77 (3%)
Frame = -2
Query: 347 SSILPACSQLKDLKSGKT-IHGFAVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMP 405
+++L AC+ + G+ + +G+ D+ + L++LYA +R+A+ + + MP
Sbjct: 780 TAVLTACNHAGFIDKGEEYFNKMITNYGLSPDIDIYACLIDLYARNGNLRKARDLMEEMP 601
Query: 406 H-RNVVTWNS-LSSCYV 420
+ N + W+S LS+C +
Sbjct: 600 YDPNCIIWSSFLSACKI 550
Score = 33.1 bits (74), Expect = 0.52
Identities = 22/109 (20%), Positives = 56/109 (51%), Gaps = 3/109 (2%)
Frame = -2
Query: 145 SSILPGCSDLQDLKSGKE-IHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMP 203
+++L C+ + G+E + + +G+ D+ + + ++ YA+ +R+A+ + + MP
Sbjct: 780 TAVLTACNHAGFIDKGEEYFNKMITNYGLSPDIDIYACLIDLYARNGNLRKARDLMEEMP 601
Query: 204 H-RDVVTWNS-LSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILS 250
+ + + W+S LS+C + + + RE + +K +P + L+
Sbjct: 600 YDPNCIIWSSFLSACKI-----YGDVELGREAAIQLIKMEPCNAAPYLT 469
Score = 30.8 bits (68), Expect = 2.6
Identities = 23/93 (24%), Positives = 48/93 (50%), Gaps = 5/93 (5%)
Frame = -2
Query: 585 SILRACSLSECLRMGKE-IHCYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPI 643
++L AC+ + + G+E + + + D+ L+D+YA+ G L +R++ + MP
Sbjct: 777 AVLTACNHAGFIDKGEEYFNKMITNYGLSPDIDIYACLIDLYARNGNLRKARDLMEEMPY 598
Query: 644 -KDVFSWNTMIFANGMHGN---GKEALSLFEKM 672
+ W++ + A ++G+ G+EA KM
Sbjct: 597 DPNCIIWSSFLSACKIYGDVELGREAAIQLIKM 499
Score = 26.2 bits (56), Expect(2) = 1e-29
Identities = 15/53 (28%), Positives = 27/53 (50%)
Frame = -3
Query: 821 SWFHVGNRVHTFVAGDKSNMESDKIYNFLDELFAKIKAAGYKPDTDYVLHDID 873
SW V H F D ++ +S++IY + ++ +I + P YV+ DI+
Sbjct: 365 SWVEVDKHNHVFAVDDVTHQQSNEIYAE*ESIYFEI--LEFSP---YVVEDIN 222
>BF640739 weakly similar to GP|7363286|dbj Similar to Arabidopsis thaliana
DNA chromosome 4 BAC clone F4I10; vegetative storage
protein., partial (14%)
Length = 444
Score = 119 bits (299), Expect = 4e-27
Identities = 65/148 (43%), Positives = 85/148 (56%), Gaps = 1/148 (0%)
Frame = +1
Query: 561 IEEAMEIFRKMQTMGFKPDETTIYSILRACSLSECLRMGKEIHCYVFRHWKDWDLARTNA 620
+ EA+ +F MQ+ G KPD T S++ A + R K IH R D ++ A
Sbjct: 1 VNEALNLFCTMQSQGIKPDSFTFVSVITALADLSVTRQAKWIHGLAIRTNMDTNVFVATA 180
Query: 621 LVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGKEALSLFEKML-LSMVKP 679
LVDMYAKCG + +R +FDMM + V +WN MI G HG GK AL LF+ M + +KP
Sbjct: 181 LVDMYAKCGAIETARELFDMMQERHVITWNAMIDGYGTHGLGKAALDLFDDMQNEASLKP 360
Query: 680 DSATFTCVLSACSHSMLVEEGVQIFNSM 707
+ TF V+SACSHS VEEG+ F M
Sbjct: 361 NDITFLSVISACSHSGFVEEGLYYFKIM 444
Score = 99.8 bits (247), Expect = 5e-21
Identities = 52/146 (35%), Positives = 83/146 (56%), Gaps = 1/146 (0%)
Frame = +1
Query: 427 KGLNVFREMVLNGVKPDLVTMLSILHACSDLQDLKSGKVIHGFAVRHGMVEDVFVCNALL 486
+ LN+F M G+KPD T +S++ A +DL + K IHG A+R M +VFV AL+
Sbjct: 7 EALNLFCTMQSQGIKPDSFTFVSVITALADLSVTRQAKWIHGLAIRTNMDTNVFVATALV 186
Query: 487 SLYAKCVCVREAQVVFDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQM-NRDEVKADE 545
+YAKC + A+ +FD++ R V +WN ++ Y T+ + L +F M N +K ++
Sbjct: 187 DMYAKCGAIETARELFDMMQERHVITWNAMIDGYGTHGLGKAALDLFDDMQNEASLKPND 366
Query: 546 ITWSVVIGGCVKNSRIEEAMEIFRKM 571
IT+ VI C + +EE + F+ M
Sbjct: 367 ITFLSVISACSHSGFVEEGLYYFKIM 444
Score = 98.2 bits (243), Expect = 1e-20
Identities = 49/131 (37%), Positives = 78/131 (59%), Gaps = 1/131 (0%)
Frame = +1
Query: 326 KGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALV 385
+ LN+F M G+KPD S++ A + L + K IHG A++ M +VFV TALV
Sbjct: 7 EALNLFCTMQSQGIKPDSFTFVSVITALADLSVTRQAKWIHGLAIRTNMDTNVFVATALV 186
Query: 386 NLYANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLN-GVKPDL 444
++YA C + A+ +FD+M R+V+TWN++ Y G + L++F +M +KP+
Sbjct: 187 DMYAKCGAIETARELFDMMQERHVITWNAMIDGYGTHGLGKAALDLFDDMQNEASLKPND 366
Query: 445 VTMLSILHACS 455
+T LS++ ACS
Sbjct: 367 ITFLSVISACS 399
Score = 95.5 bits (236), Expect = 9e-20
Identities = 49/139 (35%), Positives = 80/139 (57%), Gaps = 1/139 (0%)
Frame = +1
Query: 225 KGLNVFREMVLDGVKPDPVTVSCILSACSDLQDLKSGKAIHGFALKHGMVENVFVSNALV 284
+ LN+F M G+KPD T +++A +DL + K IHG A++ M NVFV+ ALV
Sbjct: 7 EALNLFCTMQSQGIKPDSFTFVSVITALADLSVTRQAKWIHGLAIRTNMDTNVFVATALV 186
Query: 285 NLYESCLCVREAQAVFDLMPHRNVITWNSLASCYVNCGFPQKGLNVFREM-GLNGVKPDP 343
++Y C + A+ +FD+M R+VITWN++ Y G + L++F +M +KP+
Sbjct: 187 DMYAKCGAIETARELFDMMQERHVITWNAMIDGYGTHGLGKAALDLFDDMQNEASLKPND 366
Query: 344 MAMSSILPACSQLKDLKSG 362
+ S++ ACS ++ G
Sbjct: 367 ITFLSVISACSHSGFVEEG 423
Score = 89.0 bits (219), Expect = 8e-18
Identities = 45/131 (34%), Positives = 75/131 (56%), Gaps = 1/131 (0%)
Frame = +1
Query: 124 QGLNVFRKMGLNKVKANPLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFV 183
+ LN+F M +K + T S++ +DL + K IHG +R M +VFV++A V
Sbjct: 7 EALNLFCTMQSQGIKPDSFTFVSVITALADLSVTRQAKWIHGLAIRTNMDTNVFVATALV 186
Query: 184 NFYAKCLCVREAQTVFDLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLD-GVKPDP 242
+ YAKC + A+ +FD+M R V+TWN++ Y G + L++F +M + +KP+
Sbjct: 187 DMYAKCGAIETARELFDMMQERHVITWNAMIDGYGTHGLGKAALDLFDDMQNEASLKPND 366
Query: 243 VTVSCILSACS 253
+T ++SACS
Sbjct: 367 ITFLSVISACS 399
Score = 77.0 bits (188), Expect = 3e-14
Identities = 42/132 (31%), Positives = 70/132 (52%), Gaps = 1/132 (0%)
Frame = +1
Query: 22 NEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSIGNAF 81
NEA+ ++ + +++GIKPD F++V A A + K H A R + ++V + A
Sbjct: 4 NEALNLFCTMQSQGIKPDSFTFVSVITALADLSVTRQAKWIHGLAIRTNMDTNVFVATAL 183
Query: 82 IHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKM-GLNKVKAN 140
+ Y KC +E AR +FD + R V+TWN++ Y G + L++F M +K N
Sbjct: 184 VDMYAKCGAIETARELFDMMQERHVITWNAMIDGYGTHGLGKAALDLFDDMQNEASLKPN 363
Query: 141 PLTVSSILPGCS 152
+T S++ CS
Sbjct: 364 DITFLSVISACS 399
>TC90451 similar to PIR|H84508|H84508 hypothetical protein At2g13600
[imported] - Arabidopsis thaliana, partial (12%)
Length = 1362
Score = 118 bits (295), Expect = 1e-26
Identities = 78/300 (26%), Positives = 132/300 (44%), Gaps = 9/300 (3%)
Frame = +2
Query: 382 TALVNLYANCLCVREAQTVFD-------LMPHRNVVTWNSLSSCYVNCGFPQKGLNVFRE 434
T + L +N +R+ ++ L + WN++ Y PQ L ++
Sbjct: 278 TVIATLLSNTTRIRDLNQIYAHILLTRFLESNPASFNWNNIIRSYTRLESPQNALRIYVS 457
Query: 435 MVLNGVKPDLVTMLSILHACSDLQDLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCVC 494
M+ GV PD T+ +L A S ++ G+ +H + ++ G+ + + + ++LY K
Sbjct: 458 MLRAGVLPDRYTLPIVLKAVSQSFAIQLGQQVHSYGIKLGLQSNEYCESGFINLYCKAGD 637
Query: 495 VREAQVVFDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGG 554
A VFD ++ SWN + I G
Sbjct: 638 FDSAHKVFDENHEPKLGSWNAL-----------------------------------ISG 712
Query: 555 CVKNSRIEEAMEIFRKMQTMGFKPDETTIYSILRACSLSECLRMGKEIHCYVFR-HWKDW 613
+ +A+ +F M+ GF+PD T+ S++ AC L + ++H YVF+ +W
Sbjct: 713 LSQGGLAMDAIVVFVDMKRHGFEPDGITMVSVMSACGSIGDLYLALQLHKYVFQAKTNEW 892
Query: 614 D-LARTNALVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGKEALSLFEKM 672
+ +N+L+DMY KCG + L+ VF M ++V SW +MI MHG+ KEAL F M
Sbjct: 893 TVILMSNSLIDMYGKCGRMDLAYEVFATMEDRNVSSWTSMIVGYAMHGHAKEALGCFHCM 1072
Score = 105 bits (263), Expect = 6e-23
Identities = 66/233 (28%), Positives = 106/233 (45%), Gaps = 2/233 (0%)
Frame = +2
Query: 210 WNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQDLKSGKAIHGFAL 269
WN++ Y PQ L ++ M+ GV PD T+ +L A S ++ G+ +H + +
Sbjct: 389 WNNIIRSYTRLESPQNALRIYVSMLRAGVLPDRYTLPIVLKAVSQSFAIQLGQQVHSYGI 568
Query: 270 KHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASCYVNCGFPQKGLN 329
K G+ N + + +NLY A VFD + +WN+L S G +
Sbjct: 569 KLGLQSNEYCESGFINLYCKAGDFDSAHKVFDENHEPKLGSWNALISGLSQGGLAMDAIV 748
Query: 330 VFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHG--FAVKHGMVEDVFVCTALVNL 387
VF +M +G +PD + M S++ AC + DL +H F K + + +L+++
Sbjct: 749 VFVDMKRHGFEPDGITMVSVMSACGSIGDLYLALQLHKYVFQAKTNEWTVILMSNSLIDM 928
Query: 388 YANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGV 440
Y C + A VF M RNV +W S+ Y G ++ L F M GV
Sbjct: 929 YGKCGRMDLAYEVFATMEDRNVSSWTSMIVGYAMHGHAKEALGCFHCMKGVGV 1087
Score = 104 bits (259), Expect = 2e-22
Identities = 62/228 (27%), Positives = 107/228 (46%), Gaps = 2/228 (0%)
Frame = +2
Query: 311 WNSLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAV 370
WN++ Y PQ L ++ M GV PD + +L A SQ ++ G+ +H + +
Sbjct: 389 WNNIIRSYTRLESPQNALRIYVSMLRAGVLPDRYTLPIVLKAVSQSFAIQLGQQVHSYGI 568
Query: 371 KHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLN 430
K G+ + + + +NLY A VFD + +WN+L S G +
Sbjct: 569 KLGLQSNEYCESGFINLYCKAGDFDSAHKVFDENHEPKLGSWNALISGLSQGGLAMDAIV 748
Query: 431 VFREMVLNGVKPDLVTMLSILHACSDLQDLKSGKVIHGFAVRHGMVE--DVFVCNALLSL 488
VF +M +G +PD +TM+S++ AC + DL +H + + E + + N+L+ +
Sbjct: 749 VFVDMKRHGFEPDGITMVSVMSACGSIGDLYLALQLHKYVFQAKTNEWTVILMSNSLIDM 928
Query: 489 YAKCVCVREAQVVFDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQM 536
Y KC + A VF + R V+SW ++ Y + ++ L F M
Sbjct: 929 YGKCGRMDLAYEVFATMEDRNVSSWTSMIVGYAMHGHAKEALGCFHCM 1072
Score = 99.8 bits (247), Expect = 5e-21
Identities = 64/233 (27%), Positives = 106/233 (45%), Gaps = 2/233 (0%)
Frame = +2
Query: 109 WNSLSACYVNCGFPQQGLNVFRKMGLNKVKANPLTVSSILPGCSDLQDLKSGKEIHGFVV 168
WN++ Y PQ L ++ M V + T+ +L S ++ G+++H + +
Sbjct: 389 WNNIIRSYTRLESPQNALRIYVSMLRAGVLPDRYTLPIVLKAVSQSFAIQLGQQVHSYGI 568
Query: 169 RHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVTWNSLSSCYVNCGFPQKGLN 228
+ G+ + + S F+N Y K A VFD + +WN+L S G +
Sbjct: 569 KLGLQSNEYCESGFINLYCKAGDFDSAHKVFDENHEPKLGSWNALISGLSQGGLAMDAIV 748
Query: 229 VFREMVLDGVKPDPVTVSCILSACSDLQDLKSGKAIHG--FALKHGMVENVFVSNALVNL 286
VF +M G +PD +T+ ++SAC + DL +H F K + +SN+L+++
Sbjct: 749 VFVDMKRHGFEPDGITMVSVMSACGSIGDLYLALQLHKYVFQAKTNEWTVILMSNSLIDM 928
Query: 287 YESCLCVREAQAVFDLMPHRNVITWNSLASCYVNCGFPQKGLNVFREMGLNGV 339
Y C + A VF M RNV +W S+ Y G ++ L F M GV
Sbjct: 929 YGKCGRMDLAYEVFATMEDRNVSSWTSMIVGYAMHGHAKEALGCFHCMKGVGV 1087
Score = 84.0 bits (206), Expect(2) = 6e-18
Identities = 58/220 (26%), Positives = 101/220 (45%), Gaps = 2/220 (0%)
Frame = +2
Query: 21 PNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSIGNA 80
P A++IY S G+ PD+ V KA + S +Q H + G+ S+ +
Sbjct: 428 PQNALRIYVSMLRAGVLPDRYTLPIVLKAVSQSFAIQLGQQVHSYGIKLGLQSNEYCESG 607
Query: 81 FIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVKAN 140
FI+ Y K + A +VFD+ + +WN+L + G + VF M + + +
Sbjct: 608 FINLYCKAGDFDSAHKVFDENHEPKLGSWNALISGLSQGGLAMDAIVVFVDMKRHGFEPD 787
Query: 141 PLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVE--DVFVSSAFVNFYAKCLCVREAQTV 198
+T+ S++ C + DL ++H +V + E + +S++ ++ Y KC + A V
Sbjct: 788 GITMVSVMSACGSIGDLYLALQLHKYVFQAKTNEWTVILMSNSLIDMYGKCGRMDLAYEV 967
Query: 199 FDLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGV 238
F M R+V +W S+ Y G ++ L F M GV
Sbjct: 968 FATMEDRNVSSWTSMIVGYAMHGHAKEALGCFHCMKGVGV 1087
Score = 73.9 bits (180), Expect = 3e-13
Identities = 39/98 (39%), Positives = 56/98 (56%)
Frame = +1
Query: 662 GKEALSLFEKMLLSMVKPDSATFTCVLSACSHSMLVEEGVQIFNSMSRDHLVEPEAEHYT 721
G LSL+E VKP+ TF VLSAC H V+EG F+ M + + P+ +HY
Sbjct: 1048 GTWLLSLYEGS--RGVKPNYVTFIGVLSACVHGGTVQEGRFYFDMMKNIYGITPQLQHYG 1221
Query: 722 CVVDIYSRAGCLEEAYGFIQRMPMEPTAIAWKAFLAGC 759
C+VD+ RAG ++A ++ MPM+P ++ W F GC
Sbjct: 1222 CMVDLLGRAGLFDDARRMVEEMPMKPNSVVWGVF-DGC 1332
Score = 71.2 bits (173), Expect = 2e-12
Identities = 72/332 (21%), Positives = 133/332 (39%), Gaps = 17/332 (5%)
Frame = +2
Query: 454 CSDLQDLKSGKVIHGFAVRHG--MVE--DVFVCNALLSLYAKCVCVREAQVVFDLIPHRE 509
CS L LK G VI G M+E +V C A S ++C C +P
Sbjct: 104 CS-LNCLKEGVVIVTCHATGGESMIEATNVGSCPASSSHGSRCFCS---------VPQHS 253
Query: 510 VASWNGILTAYFTNKEYEKGLYMFSQMNRD-------EVKADEITWSVVIGGCVKNSRIE 562
+ S N +T T + +Q+ E W+ +I + +
Sbjct: 254 ITSGNDPVTVIATLLSNTTRIRDLNQIYAHILLTRFLESNPASFNWNNIIRSYTRLESPQ 433
Query: 563 EAMEIFRKMQTMGFKPDETTIYSILRACSLSECLRMGKEIHCYVFRHWKDWDLARTNALV 622
A+ I+ M G PD T+ +L+A S S +++G+++H Y + + + +
Sbjct: 434 NALRIYVSMLRAGVLPDRYTLPIVLKAVSQSFAIQLGQQVHSYGIKLGLQSNEYCESGFI 613
Query: 623 DMYAKCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGKEALSLFEKMLLSMVKPDSA 682
++Y K G + VFD + SWN +I G +A+ +F M +PD
Sbjct: 614 NLYCKAGDFDSAHKVFDENHEPKLGSWNALISGLSQGGLAMDAIVVFVDMKRHGFEPDGI 793
Query: 683 TFTCVLSACSHSMLVEEGVQIFNSMSRDHLVEPEAEHYTCV------VDIYSRAGCLEEA 736
T V+SAC + +Q+ ++ + + +T + +D+Y + G ++ A
Sbjct: 794 TMVSVMSACGSIGDLYLALQL-----HKYVFQAKTNEWTVILMSNSLIDMYGKCGRMDLA 958
Query: 737 YGFIQRMPMEPTAIAWKAFLAGCRVYKNVELA 768
Y M + +W + + G ++ + + A
Sbjct: 959 YEVFATME-DRNVSSWTSMIVGYAMHGHAKEA 1051
Score = 52.0 bits (123), Expect = 1e-06
Identities = 31/116 (26%), Positives = 52/116 (44%), Gaps = 2/116 (1%)
Frame = +2
Query: 19 GLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDA--TRCGVMSDVS 76
GL +AI ++ + G +PD ++V AC + D Q H + + +
Sbjct: 725 GLAMDAIVVFVDMKRHGFEPDGITMVSVMSACGSIGDLYLALQLHKYVFQAKTNEWTVIL 904
Query: 77 IGNAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKM 132
+ N+ I YGKC ++ A VF + R+V +W S+ Y G ++ L F M
Sbjct: 905 MSNSLIDMYGKCGRMDLAYEVFATMEDRNVSSWTSMIVGYAMHGHAKEALGCFHCM 1072
Score = 25.8 bits (55), Expect(2) = 6e-18
Identities = 20/83 (24%), Positives = 36/83 (43%), Gaps = 2/83 (2%)
Frame = +1
Query: 237 GVKPDPVTVSCILSACSDLQDLKSGKAIHGFALK-HGMVENVFVSNALVNLYESCLCVRE 295
GVKP+ VT +LSAC ++ G+ +G+ + +V+L +
Sbjct: 1084 GVKPNYVTFIGVLSACVHGGTVQEGRFYFDMMKNIYGITPQLQHYGCMVDLLGRAGLFDD 1263
Query: 296 AQAVFDLMPHR-NVITWNSLASC 317
A+ + + MP + N + W C
Sbjct: 1264 ARRMVEEMPMKPNSVVWGVFDGC 1332
>BG646586 weakly similar to GP|10177683|dbj selenium-binding protein-like
{Arabidopsis thaliana}, partial (9%)
Length = 687
Score = 108 bits (270), Expect = 1e-23
Identities = 52/110 (47%), Positives = 68/110 (61%)
Frame = -3
Query: 713 VEPEAEHYTCVVDIYSRAGCLEEAYGFIQRMPMEPTAIAWKAFLAGCRVYKNVELAKISA 772
+EP EHYT +VD+ RAG LE+AY I+ A W + LA CRVY NVEL +I+A
Sbjct: 664 IEPLPEHYTGMVDLLGRAGQLEKAYSLIKENSTSADATTWGSLLAACRVYGNVELGEIAA 485
Query: 773 KKLFEIDPNGSANYVTLFNILVTAKLWSEASKIRKLMKERGITKTPGCSW 822
+ LFEIDP S NYV L N + W A +++KLM ++G+ K G SW
Sbjct: 484 RHLFEIDPTDSGNYVLLANTYASNDKWERAEEVKKLMSKKGMKKPSGYSW 335
>BE202782 weakly similar to GP|22093801|dbj hypothetical protein~predicted by
GlimmerM~similar to Arabidopsis thaliana chromosome3
At3g23330, partial (13%)
Length = 548
Score = 108 bits (270), Expect = 1e-23
Identities = 49/93 (52%), Positives = 63/93 (67%)
Frame = +2
Query: 860 GYKPDTDYVLHDIDQEEKAESLCNHSEKLAVAFGILNLNGQSTIRVFKNLRICGDCHNAI 919
G P T VL D+++++K + LC HSEKLAV G++N + ++V KNLRIC DCH I
Sbjct: 2 GCLPMTKSVLQDVEEQDKEQILCGHSEKLAVVLGLINTSPGQPLQVIKNLRICDDCHAVI 181
Query: 920 KYMSNVVGVTIVVRDSLRFHHFKNGNCSCKDFW 952
K +S + G I VRD+ RFHHFK G CSC DFW
Sbjct: 182 KVISRLEGREIFVRDTNRFHHFKEGVCSCADFW 280
>BF642497 weakly similar to PIR|H96713|H96 hypothetical protein T6L1.11
[imported] - Arabidopsis thaliana, partial (20%)
Length = 461
Score = 107 bits (266), Expect = 3e-23
Identities = 44/152 (28%), Positives = 90/152 (58%)
Frame = +3
Query: 208 VTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQDLKSGKAIHGF 267
++W S+ + + G + +++FREM L+ ++ D T +L+AC + L+ GK +H +
Sbjct: 6 ISWTSMITGFTQNGLDRDAIDIFREMKLENLQMDQYTFGSVLTACGGVMALQEGKQVHAY 185
Query: 268 ALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASCYVNCGFPQKG 327
++ +N+FV++ALV++Y C ++ A+AVF M +NV++W ++ Y G+ ++
Sbjct: 186 IIRTDYKDNIFVASALVDMYCKCKNIKSAEAVFKKMTCKNVVSWTAMLVGYGQNGYSEEA 365
Query: 328 LNVFREMGLNGVKPDPMAMSSILPACSQLKDL 359
+ F +M G++PD + S++ +C+ L L
Sbjct: 366 VKTFSDMQKYGIEPDDFTLGSVISSCANLASL 461
Score = 105 bits (261), Expect = 1e-22
Identities = 43/152 (28%), Positives = 89/152 (58%)
Frame = +3
Query: 107 VTWNSLSACYVNCGFPQQGLNVFRKMGLNKVKANPLTVSSILPGCSDLQDLKSGKEIHGF 166
++W S+ + G + +++FR+M L ++ + T S+L C + L+ GK++H +
Sbjct: 6 ISWTSMITGFTQNGLDRDAIDIFREMKLENLQMDQYTFGSVLTACGGVMALQEGKQVHAY 185
Query: 167 VVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVTWNSLSSCYVNCGFPQKG 226
++R +++FV+SA V+ Y KC ++ A+ VF M ++VV+W ++ Y G+ ++
Sbjct: 186 IIRTDYKDNIFVASALVDMYCKCKNIKSAEAVFKKMTCKNVVSWTAMLVGYGQNGYSEEA 365
Query: 227 LNVFREMVLDGVKPDPVTVSCILSACSDLQDL 258
+ F +M G++PD T+ ++S+C++L L
Sbjct: 366 VKTFSDMQKYGIEPDDFTLGSVISSCANLASL 461
Score = 103 bits (256), Expect = 4e-22
Identities = 45/152 (29%), Positives = 88/152 (57%)
Frame = +3
Query: 309 ITWNSLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGF 368
I+W S+ + + G + +++FREM L ++ D S+L AC + L+ GK +H +
Sbjct: 6 ISWTSMITGFTQNGLDRDAIDIFREMKLENLQMDQYTFGSVLTACGGVMALQEGKQVHAY 185
Query: 369 AVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKG 428
++ +++FV +ALV++Y C ++ A+ VF M +NVV+W ++ Y G+ ++
Sbjct: 186 IIRTDYKDNIFVASALVDMYCKCKNIKSAEAVFKKMTCKNVVSWTAMLVGYGQNGYSEEA 365
Query: 429 LNVFREMVLNGVKPDLVTMLSILHACSDLQDL 460
+ F +M G++PD T+ S++ +C++L L
Sbjct: 366 VKTFSDMQKYGIEPDDFTLGSVISSCANLASL 461
Score = 98.6 bits (244), Expect = 1e-20
Identities = 52/149 (34%), Positives = 85/149 (56%), Gaps = 1/149 (0%)
Frame = +3
Query: 546 ITWSVVIGGCVKNSRIEEAMEIFRKMQTMGFKPDETTIYSILRACSLSECLRMGKEIHCY 605
I+W+ +I G +N +A++IFR+M+ + D+ T S+L AC L+ GK++H Y
Sbjct: 6 ISWTSMITGFTQNGLDRDAIDIFREMKLENLQMDQYTFGSVLTACGGVMALQEGKQVHAY 185
Query: 606 VFR-HWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGKE 664
+ R +KD ++ +ALVDMY KC + + VF M K+V SW M+ G +G +E
Sbjct: 186 IIRTDYKD-NIFVASALVDMYCKCKNIKSAEAVFKKMTCKNVVSWTAMLVGYGQNGYSEE 362
Query: 665 ALSLFEKMLLSMVKPDSATFTCVLSACSH 693
A+ F M ++PD T V+S+C++
Sbjct: 363 AVKTFSDMQKYGIEPDDFTLGSVISSCAN 449
Score = 95.9 bits (237), Expect = 7e-20
Identities = 44/146 (30%), Positives = 79/146 (53%)
Frame = +3
Query: 410 VTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHACSDLQDLKSGKVIHGF 469
++W S+ + + G + +++FREM L ++ D T S+L AC + L+ GK +H +
Sbjct: 6 ISWTSMITGFTQNGLDRDAIDIFREMKLENLQMDQYTFGSVLTACGGVMALQEGKQVHAY 185
Query: 470 AVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHREVASWNGILTAYFTNKEYEKG 529
+R +++FV +AL+ +Y KC ++ A+ VF + + V SW +L Y N E+
Sbjct: 186 IIRTDYKDNIFVASALVDMYCKCKNIKSAEAVFKKMTCKNVVSWTAMLVGYGQNGYSEEA 365
Query: 530 LYMFSQMNRDEVKADEITWSVVIGGC 555
+ FS M + ++ D+ T VI C
Sbjct: 366 VKTFSDMQKYGIEPDDFTLGSVISSC 443
Score = 69.7 bits (169), Expect = 5e-12
Identities = 34/140 (24%), Positives = 68/140 (48%)
Frame = +3
Query: 18 HGLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSI 77
+GL +AI I+ + ++ D+ F +V AC + KQ H R ++ +
Sbjct: 42 NGLDRDAIDIFREMKLENLQMDQYTFGSVLTACGGVMALQEGKQVHAYIIRTDYKDNIFV 221
Query: 78 GNAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKV 137
+A + Y KCK ++ A VF + ++VV+W ++ Y G+ ++ + F M +
Sbjct: 222 ASALVDMYCKCKNIKSAEAVFKKMTCKNVVSWTAMLVGYGQNGYSEEAVKTFSDMQKYGI 401
Query: 138 KANPLTVSSILPGCSDLQDL 157
+ + T+ S++ C++L L
Sbjct: 402 EPDDFTLGSVISSCANLASL 461
Score = 50.1 bits (118), Expect = 4e-06
Identities = 32/115 (27%), Positives = 58/115 (49%), Gaps = 4/115 (3%)
Frame = +3
Query: 648 SWNTMIFANGMHGNGKEALSLFEKMLLSMVKPDSATFTCVLSACSHSMLVEEGVQ----I 703
SW +MI +G ++A+ +F +M L ++ D TF VL+AC M ++EG Q I
Sbjct: 9 SWTSMITGFTQNGLDRDAIDIFREMKLENLQMDQYTFGSVLTACGGVMALQEGKQVHAYI 188
Query: 704 FNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAYGFIQRMPMEPTAIAWKAFLAG 758
+ +D++ A +VD+Y + ++ A ++M + ++W A L G
Sbjct: 189 IRTDYKDNIFVASA-----LVDMYCKCKNIKSAEAVFKKMTCK-NVVSWTAMLVG 335
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.324 0.137 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,715,810
Number of Sequences: 36976
Number of extensions: 530065
Number of successful extensions: 3558
Number of sequences better than 10.0: 124
Number of HSP's better than 10.0 without gapping: 1968
Number of HSP's successfully gapped in prelim test: 68
Number of HSP's that attempted gapping in prelim test: 634
Number of HSP's gapped (non-prelim): 2880
length of query: 952
length of database: 9,014,727
effective HSP length: 105
effective length of query: 847
effective length of database: 5,132,247
effective search space: 4347013209
effective search space used: 4347013209
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 63 (28.9 bits)
Medicago: description of AC124970.5


