

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124968.5 - phase: 0
(732 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
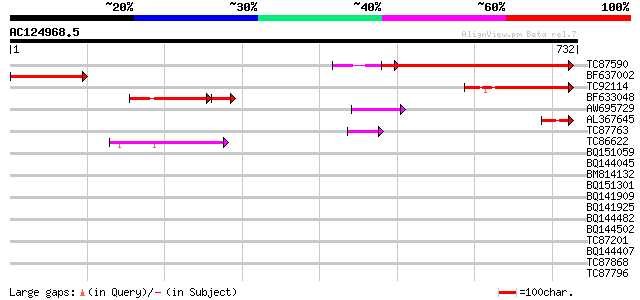
Score E
Sequences producing significant alignments: (bits) Value
TC87590 weakly similar to PIR|G96522|G96522 F11A17.16 [imported]... 238 6e-63
BF637002 210 2e-54
TC92114 similar to GP|11994760|dbj|BAB03089. contains similarity... 135 7e-32
BF633048 similar to GP|11994760|d contains similarity to pheroph... 61 4e-15
AW695729 similar to GP|22135814|gb| AT4g18560/F28J12_220 {Arabid... 47 3e-05
AL367645 similar to GP|22135814|gb| AT4g18560/F28J12_220 {Arabid... 45 8e-05
TC87763 similar to GP|21593475|gb|AAM65442.1 unknown {Arabidopsi... 44 2e-04
TC86622 weakly similar to GP|20197623|gb|AAM15156.1 putative myo... 43 4e-04
BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melano... 42 0.001
BQ144045 similar to PIR|T51947|T51 probable transcription factor... 40 0.003
BM814132 weakly similar to OMNI|MT3615.1 PE_PGRS family protein ... 39 0.007
BQ151301 similar to PIR|S34666|S346 glycine-rich protein - commo... 39 0.007
BQ141909 weakly similar to GP|14253159|emb VMP3 protein {Volvox ... 39 0.007
BQ141925 homologue to GP|10727150|gb| Rm62 gene product {alt 1} ... 39 0.007
BQ144482 similar to GP|6523547|emb| hydroxyproline-rich glycopro... 39 0.009
BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 39 0.009
TC87201 similar to PIR|F86388|F86388 hypothetical protein AAG292... 39 0.009
BQ144407 weakly similar to GP|6523547|emb| hydroxyproline-rich g... 38 0.012
TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 38 0.012
TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear prot... 38 0.012
>TC87590 weakly similar to PIR|G96522|G96522 F11A17.16 [imported] -
Arabidopsis thaliana, partial (45%)
Length = 2425
Score = 238 bits (607), Expect = 6e-63
Identities = 119/248 (47%), Positives = 168/248 (66%)
Frame = +1
Query: 481 NTAMVKRAPQVVELYHSLMKRDSRRDSSSGGLSDAPDVADVRSSMIGEIENRSSHLLAIK 540
N +KR Q+ + R ++D P +S++GEI+NRS+HLLAI+
Sbjct: 1135 NWLTLKRLLQLSNYFTP*RIRTPKKDLKGSINHQKPITNSAHNSIVGEIQNRSAHLLAIR 1314
Query: 541 ADIETQGEFVNSLIREVNDAVYENIDDVVAFVKWLDDELGFLVDERAVLKHFDWPEKKAD 600
DI+T+GEF+N LI +V DA Y +I+DV+ FV WLD EL L DERAVLKHF WPE+KAD
Sbjct: 1315 EDIQTKGEFINGLINKVVDASYVDIEDVLKFVDWLDGELSTLADERAVLKHFKWPERKAD 1494
Query: 601 TLREAAFGYQDLKKLESEVSSYKDDPRLPCDIALKKMVALSEKMERTVYTLLRTRDSLMR 660
T+REAA Y++LK LE E+SSYKDDP +PC +LKK+ +L +K ER++ L+ R+S++R
Sbjct: 1495 TMREAAVEYRELKMLEQEISSYKDDPDIPCVASLKKIASLLDKSERSIQKLIVLRNSVIR 1674
Query: 661 NCKEFQIPVEWMLDNGIIGKIKLGSVKLAKKYMKRVAIEVQTKSAFDKDPAMDYMVLQGV 720
+ + + IP WMLD+GI KIK S+ L K YMKR+ +E+++ D++ D ++LQGV
Sbjct: 1675 SYQMYNIPTAWMLDSGISSKIKQSSMTLVKMYMKRLTMELESIRNSDRESNQDSLLLQGV 1854
Query: 721 RFAFRIHQ 728
FA+R HQ
Sbjct: 1855 HFAYRAHQ 1878
Score = 47.4 bits (111), Expect = 2e-05
Identities = 29/87 (33%), Positives = 40/87 (45%)
Frame = +3
Query: 417 KESVPMPLFVQQCALEKRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSF 476
K S+P P+ E +L +PP + PPPPPPPP P
Sbjct: 993 KSSIPAPIPNHAAIREITSLGRKSPPNH--------------CLMPPPPPPPPPIPSRPL 1130
Query: 477 ASRGNTAMVKRAPQVVELYHSLMKRDS 503
A NT ++AP VV+L+HSL +D+
Sbjct: 1131 AKLANT---QKAPAVVQLFHSLKNQDT 1202
>BF637002
Length = 378
Score = 210 bits (534), Expect = 2e-54
Identities = 100/100 (100%), Positives = 100/100 (100%)
Frame = +2
Query: 1 MKEEIHNNPSENKTKVSKFSDQNQPPKLQTTKTTNPNNNNHSKPRLWGAHIVKGFSADKK 60
MKEEIHNNPSENKTKVSKFSDQNQPPKLQTTKTTNPNNNNHSKPRLWGAHIVKGFSADKK
Sbjct: 77 MKEEIHNNPSENKTKVSKFSDQNQPPKLQTTKTTNPNNNNHSKPRLWGAHIVKGFSADKK 256
Query: 61 TKQLPTKKQQNTTTITTSDVVTNQKNVNPFVPPHSRVKRS 100
TKQLPTKKQQNTTTITTSDVVTNQKNVNPFVPPHSRVKRS
Sbjct: 257 TKQLPTKKQQNTTTITTSDVVTNQKNVNPFVPPHSRVKRS 376
>TC92114 similar to GP|11994760|dbj|BAB03089. contains similarity to
pherophorin~gene_id:T5M7.14 {Arabidopsis thaliana},
partial (15%)
Length = 674
Score = 135 bits (339), Expect = 7e-32
Identities = 72/146 (49%), Positives = 100/146 (68%), Gaps = 5/146 (3%)
Frame = +3
Query: 588 VLKHFDWPEKKADTLREAAFGYQDLK-----KLESEVSSYKDDPRLPCDIALKKMVALSE 642
VLKHFDWPE KAD + + ++K ++ES SS P + LKKM +L E
Sbjct: 3 VLKHFDWPEGKADAPKGGSL--LNIKI**NWRIESLPSSMI--PNSHVKLRLKKMYSLLE 170
Query: 643 KMERTVYTLLRTRDSLMRNCKEFQIPVEWMLDNGIIGKIKLGSVKLAKKYMKRVAIEVQT 702
K+E++VY LLRT+D + +EF IP+ W+ D G++GKIKL SV+LA+KYMKRVA E+
Sbjct: 171 KVEQSVYALLRTKDMAISRYREFGIPINWLQDAGVVGKIKLSSVQLARKYMKRVASELDA 350
Query: 703 KSAFDKDPAMDYMVLQGVRFAFRIHQ 728
S +K+PA ++++LQGVRFAFR+HQ
Sbjct: 351 LSGPEKEPAREFLILQGVRFAFRVHQ 428
>BF633048 similar to GP|11994760|d contains similarity to
pherophorin~gene_id:T5M7.14 {Arabidopsis thaliana},
partial (15%)
Length = 510
Score = 60.8 bits (146), Expect(2) = 4e-15
Identities = 40/107 (37%), Positives = 65/107 (60%), Gaps = 1/107 (0%)
Frame = +2
Query: 155 RKNQSEVDELVKKVALLEEEKSGLSEQLVALSRSCGLERQEEDKDGSTQN-LELEVVELR 213
R+ Q E ++ ++ LL+++ SGL + + ++ E DK N LE+ VVEL+
Sbjct: 107 RQMQLEANQTKGQLLLLKQQVSGLQVK----EEAGAIKDAEIDKKLKAVNDLEVAVVELK 274
Query: 214 RLNKELHMQKRNLTCRLSSMESQLSCSDNSSESDIVAKFKAEASLLR 260
R NKEL +KR LT +L++ ES+++ N +E+++VAK K E S LR
Sbjct: 275 RKNKELQYEKRELTVKLNAAESRVAELSNMTETEMVAKAKEEVSNLR 415
Score = 38.9 bits (89), Expect(2) = 4e-15
Identities = 19/31 (61%), Positives = 23/31 (73%)
Frame = +3
Query: 261 LTNEDLSKQVEGLQTSRLNEVEELAYLRWVN 291
+ NEDLSKQVEGLQ +R +E+EEL VN
Sbjct: 417 MQNEDLSKQVEGLQMNRFSEIEELDTFVGVN 509
>AW695729 similar to GP|22135814|gb| AT4g18560/F28J12_220 {Arabidopsis
thaliana}, partial (7%)
Length = 408
Score = 47.0 bits (110), Expect = 3e-05
Identities = 28/72 (38%), Positives = 38/72 (51%), Gaps = 3/72 (4%)
Frame = +3
Query: 442 PPRPSCSISSKTKQECSAQVQPPPPPP-PPPPPMSFASRGNTAMVKRAPQVVELYHSLMK 500
P S ++ K K P P PPP + + A V++ P+VVE YHSLM+
Sbjct: 177 PFHSSLMVAGKVKVAMGFSKSPSPAHAHSTPPPPAKGGKMPPAKVRKVPEVVEFYHSLMR 356
Query: 501 RDS--RRDSSSG 510
RDS RR+S+SG
Sbjct: 357 RDSQTRRESNSG 392
>AL367645 similar to GP|22135814|gb| AT4g18560/F28J12_220 {Arabidopsis
thaliana}, partial (10%)
Length = 522
Score = 45.4 bits (106), Expect = 8e-05
Identities = 22/42 (52%), Positives = 30/42 (71%)
Frame = +1
Query: 687 KLAKKYMKRVAIEVQTKSAFDKDPAMDYMVLQGVRFAFRIHQ 728
KLA KYMKRV+ E++T P + +++QGVRFAFR+HQ
Sbjct: 1 KLAMKYMKRVSAELETVGG---GPEEEELIVQGVRFAFRVHQ 117
>TC87763 similar to GP|21593475|gb|AAM65442.1 unknown {Arabidopsis
thaliana}, partial (52%)
Length = 840
Score = 43.9 bits (102), Expect = 2e-04
Identities = 20/46 (43%), Positives = 25/46 (53%)
Frame = -1
Query: 437 RIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFASRGNT 482
R+ NPPP P+ ++ K+E QPPP P PPP P S G T
Sbjct: 591 RVSNPPPTPTATLGPGAKKENGDHPQPPPYPYPPPYPYPPTSGGAT 454
>TC86622 weakly similar to GP|20197623|gb|AAM15156.1 putative myosin heavy
chain {Arabidopsis thaliana}, partial (17%)
Length = 2546
Score = 43.1 bits (100), Expect = 4e-04
Identities = 45/169 (26%), Positives = 76/169 (44%), Gaps = 15/169 (8%)
Frame = +2
Query: 129 ELDHMRSLLQES------KEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQL 182
EL R+L ES ++ E KL + + SEV L++K+ +LEE +G EQ
Sbjct: 404 ELTSARNLELESLHESLTRDSEQKLQEAIEKFNSKDSEVQSLLEKIKILEENIAGAGEQS 583
Query: 183 VAL------SRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQ 236
++L S S Q E++D Q +E E + ++ + N+ + E Q
Sbjct: 584 ISLKSEFEESLSKLASLQSENEDLKRQIVEAEKKTSQSFSENELLVGTNIQLKTKIDELQ 763
Query: 237 LSCSDNSSESDIVA-KFKAEASLLRLTNEDLSK--QVEGLQTSRLNEVE 282
S + SE ++ A + + +LL N+ SK ++ R+ EVE
Sbjct: 764 ESLNSVVSEKEVTAQELVSHKNLLAELNDVQSKSSEIHSANEVRILEVE 910
>BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melanogaster},
partial (26%)
Length = 308
Score = 41.6 bits (96), Expect = 0.001
Identities = 18/46 (39%), Positives = 22/46 (47%)
Frame = -2
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFASRGNTAM 484
P PPP P PPPPPPPPPPP F +G++ +
Sbjct: 139 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPGVFPGKGDSCL 2
Score = 38.9 bits (89), Expect = 0.007
Identities = 17/39 (43%), Positives = 18/39 (45%)
Frame = -3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFA 477
P PPP P PPPPPPPPPPP F+
Sbjct: 141 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPGFS 25
Score = 37.0 bits (84), Expect = 0.027
Identities = 16/35 (45%), Positives = 16/35 (45%)
Frame = -3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPPPPP
Sbjct: 255 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 151
Score = 37.0 bits (84), Expect = 0.027
Identities = 16/35 (45%), Positives = 16/35 (45%)
Frame = -3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPPPPP
Sbjct: 231 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 127
Score = 37.0 bits (84), Expect = 0.027
Identities = 16/35 (45%), Positives = 16/35 (45%)
Frame = -3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPPPPP
Sbjct: 246 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 142
Score = 37.0 bits (84), Expect = 0.027
Identities = 16/35 (45%), Positives = 16/35 (45%)
Frame = -3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPPPPP
Sbjct: 243 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 139
Score = 37.0 bits (84), Expect = 0.027
Identities = 16/35 (45%), Positives = 16/35 (45%)
Frame = -3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPPPPP
Sbjct: 237 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 133
Score = 37.0 bits (84), Expect = 0.027
Identities = 16/35 (45%), Positives = 16/35 (45%)
Frame = -1
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPPPPP
Sbjct: 215 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 111
Score = 37.0 bits (84), Expect = 0.027
Identities = 16/35 (45%), Positives = 16/35 (45%)
Frame = -3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPPPPP
Sbjct: 291 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 187
Score = 37.0 bits (84), Expect = 0.027
Identities = 16/35 (45%), Positives = 16/35 (45%)
Frame = -3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPPPPP
Sbjct: 297 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 193
Score = 37.0 bits (84), Expect = 0.027
Identities = 16/35 (45%), Positives = 16/35 (45%)
Frame = -3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPPPPP
Sbjct: 228 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 124
Score = 37.0 bits (84), Expect = 0.027
Identities = 16/35 (45%), Positives = 16/35 (45%)
Frame = -2
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPPPPP
Sbjct: 226 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 122
Score = 37.0 bits (84), Expect = 0.027
Identities = 16/35 (45%), Positives = 16/35 (45%)
Frame = -3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPPPPP
Sbjct: 222 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 118
Score = 37.0 bits (84), Expect = 0.027
Identities = 16/35 (45%), Positives = 16/35 (45%)
Frame = -3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPPPPP
Sbjct: 219 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 115
Score = 37.0 bits (84), Expect = 0.027
Identities = 16/35 (45%), Positives = 16/35 (45%)
Frame = -3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPPPPP
Sbjct: 216 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 112
Score = 37.0 bits (84), Expect = 0.027
Identities = 16/35 (45%), Positives = 16/35 (45%)
Frame = -3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPPPPP
Sbjct: 240 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 136
Score = 37.0 bits (84), Expect = 0.027
Identities = 16/35 (45%), Positives = 16/35 (45%)
Frame = -3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPPPPP
Sbjct: 252 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 148
Score = 37.0 bits (84), Expect = 0.027
Identities = 16/35 (45%), Positives = 16/35 (45%)
Frame = -3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPPPPP
Sbjct: 249 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 145
Score = 37.0 bits (84), Expect = 0.027
Identities = 16/35 (45%), Positives = 16/35 (45%)
Frame = -3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPPPPP
Sbjct: 234 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 130
Score = 34.3 bits (77), Expect = 0.18
Identities = 11/11 (100%), Positives = 11/11 (100%)
Frame = -3
Query: 463 PPPPPPPPPPP 473
PPPPPPPPPPP
Sbjct: 300 PPPPPPPPPPP 268
Score = 34.3 bits (77), Expect = 0.18
Identities = 11/11 (100%), Positives = 11/11 (100%)
Frame = -3
Query: 463 PPPPPPPPPPP 473
PPPPPPPPPPP
Sbjct: 303 PPPPPPPPPPP 271
Score = 34.3 bits (77), Expect = 0.18
Identities = 11/11 (100%), Positives = 11/11 (100%)
Frame = -2
Query: 463 PPPPPPPPPPP 473
PPPPPPPPPPP
Sbjct: 307 PPPPPPPPPPP 275
>BQ144045 similar to PIR|T51947|T51 probable transcription factor HUA2
[imported] - Arabidopsis thaliana, partial (1%)
Length = 1172
Score = 40.0 bits (92), Expect = 0.003
Identities = 20/42 (47%), Positives = 22/42 (51%)
Frame = +3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFASRG 480
P PPPR +CS+ PPPPPPPPPPP RG
Sbjct: 9 PLPPPR-ACSLGF-----------PPPPPPPPPPPCWSVGRG 98
Score = 35.4 bits (80), Expect = 0.079
Identities = 24/66 (36%), Positives = 30/66 (45%), Gaps = 9/66 (13%)
Frame = +2
Query: 441 PPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPM--SFASRGNTAMVKRA-----PQV-- 491
PPP PS C PP PPPP PPP+ +A G V+R+ P +
Sbjct: 2 PPPAPS--------PPCLFPWVPPSPPPPAPPPLLVRWAGAGGRVAVRRSVCPHPPSLWC 157
Query: 492 VELYHS 497
V L+HS
Sbjct: 158 VSLFHS 175
>BM814132 weakly similar to OMNI|MT3615.1 PE_PGRS family protein
{Mycobacterium tuberculosis CDC1551}, partial (7%)
Length = 164
Score = 38.9 bits (89), Expect = 0.007
Identities = 17/35 (48%), Positives = 17/35 (48%)
Frame = -2
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P A PPPPPPPPPPP
Sbjct: 124 PPPPPPPPPPPPPPPPPPPPAPPPPPPPPPPPPPP 20
Score = 38.5 bits (88), Expect = 0.009
Identities = 16/35 (45%), Positives = 18/35 (50%)
Frame = -1
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P + +PPPPPPPPPPP
Sbjct: 134 PPPPPPPPPPPPAPPPPPPPPPPRPPPPPPPPPPP 30
Score = 38.1 bits (87), Expect = 0.012
Identities = 16/35 (45%), Positives = 16/35 (45%)
Frame = -2
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPPPPP
Sbjct: 118 PPPPPPPPPPPPPPPPPPAPPPPPPPPPPPPPPPP 14
Score = 37.7 bits (86), Expect = 0.016
Identities = 17/35 (48%), Positives = 17/35 (48%)
Frame = -1
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPPRP PPPPPPPPPPP
Sbjct: 80 PPPPPRPP------------PPPPPPPPPPPPPPP 12
Score = 37.7 bits (86), Expect = 0.016
Identities = 16/35 (45%), Positives = 17/35 (47%)
Frame = -2
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P + PPPPPPPPPPP
Sbjct: 121 PPPPPPPPPPPPPPPPPPPAPPPPPPPPPPPPPPP 17
Score = 37.7 bits (86), Expect = 0.016
Identities = 16/35 (45%), Positives = 16/35 (45%)
Frame = -2
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPPPPP
Sbjct: 130 PPPPPPPPPPPPPPPPPPPPPPAPPPPPPPPPPPP 26
Score = 35.4 bits (80), Expect = 0.079
Identities = 16/35 (45%), Positives = 16/35 (45%)
Frame = -2
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPPPPP
Sbjct: 142 PPPPPPP-----------------PPPPPPPPPPP 89
Score = 35.4 bits (80), Expect = 0.079
Identities = 16/35 (45%), Positives = 16/35 (45%)
Frame = -1
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPPPPP
Sbjct: 158 PPPPPPP-----------------PPPPPPPPPPP 105
Score = 35.4 bits (80), Expect = 0.079
Identities = 16/35 (45%), Positives = 16/35 (45%)
Frame = -1
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPPPPP
Sbjct: 161 PPPPPPP-----------------PPPPPPPPPPP 108
Score = 35.4 bits (80), Expect = 0.079
Identities = 16/35 (45%), Positives = 16/35 (45%)
Frame = -3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPPPPP
Sbjct: 153 PPPPPPP-----------------PPPPPPPPPPP 100
Score = 35.4 bits (80), Expect = 0.079
Identities = 16/35 (45%), Positives = 16/35 (45%)
Frame = -3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPPPPP
Sbjct: 162 PPPPPPP-----------------PPPPPPPPPPP 109
Score = 33.9 bits (76), Expect = 0.23
Identities = 15/35 (42%), Positives = 15/35 (42%)
Frame = -2
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPP PPP
Sbjct: 106 PPPPPPPPPPPPPPAPPPPPPPPPPPPPPPPRPPP 2
Score = 33.5 bits (75), Expect = 0.30
Identities = 15/35 (42%), Positives = 15/35 (42%)
Frame = -2
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPP PP
Sbjct: 109 PPPPPPPPPPPPPPPAPPPPPPPPPPPPPPPPRPP 5
Score = 32.0 bits (71), Expect = 0.87
Identities = 15/35 (42%), Positives = 15/35 (42%)
Frame = -3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPP PP
Sbjct: 144 PPPPPPP-----------------PPPPPPPPRPP 91
Score = 32.0 bits (71), Expect = 0.87
Identities = 15/35 (42%), Positives = 15/35 (42%)
Frame = -3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P PPPPPPPPP P
Sbjct: 147 PPPPPPP-----------------PPPPPPPPPRP 94
>BQ151301 similar to PIR|S34666|S346 glycine-rich protein - common tobacco,
partial (22%)
Length = 530
Score = 38.9 bits (89), Expect = 0.007
Identities = 17/41 (41%), Positives = 22/41 (53%)
Frame = +2
Query: 432 EKRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPP 472
+K+ + P+ PP P E A + PPPPPPPPPP
Sbjct: 92 QKQKKKKPHHPPPPP-------PHEAGAVIMPPPPPPPPPP 193
Score = 35.8 bits (81), Expect = 0.060
Identities = 21/50 (42%), Positives = 23/50 (46%), Gaps = 9/50 (18%)
Frame = +1
Query: 433 KRALRIPNPPPRPSCSISSKTKQECSAQVQPPPP---------PPPPPPP 473
KR I PPP P S+ + E Q PPPP PPPPPPP
Sbjct: 4 KRIYFIFPPPPLPQ----SRGRGEEKTQTHPPPPKTKKKKTTPPPPPPPP 141
Score = 35.4 bits (80), Expect = 0.079
Identities = 18/44 (40%), Positives = 22/44 (49%), Gaps = 8/44 (18%)
Frame = +3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPP--------PPPPPPPPM 474
PNPPP P +K K+ + PPP PP PPPPP+
Sbjct: 72 PNPPPPPK----NKKKKNHTTPPPPPPMRRGR*LCPPRPPPPPL 191
Score = 31.6 bits (70), Expect = 1.1
Identities = 13/32 (40%), Positives = 17/32 (52%)
Frame = +1
Query: 441 PPPRPSCSISSKTKQECSAQVQPPPPPPPPPP 472
P PR + ++E + Q PPPPPPPP
Sbjct: 271 PLPRKKGGGGRRRRKERKKRKQKTPPPPPPPP 366
Score = 31.2 bits (69), Expect = 1.5
Identities = 17/42 (40%), Positives = 18/42 (42%), Gaps = 2/42 (4%)
Frame = +3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPP--PPPPPPMSFAS 478
P PPP P PPPPP PP PPP+ F S
Sbjct: 168 PRPPPPP-----------LPHGTHPPPPPHHPPTPPPLFFFS 260
Score = 30.0 bits (66), Expect = 3.3
Identities = 17/69 (24%), Positives = 28/69 (39%)
Frame = +3
Query: 441 PPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFASRGNTAMVKRAPQVVELYHSLMK 500
P P P + ++E + + PPPPPPP + V V+ YH ++
Sbjct: 267 PTPSPEKRGGGEEEKERKKEKETKDPPPPPPPTTTIYLFIYV*WVLHYYLVISAYHYMII 446
Query: 501 RDSRRDSSS 509
R ++ S
Sbjct: 447 RIEKKTKKS 473
Score = 30.0 bits (66), Expect = 3.3
Identities = 13/42 (30%), Positives = 20/42 (46%)
Frame = +1
Query: 435 ALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSF 476
+L P P + + K+ + + PPPPPPPP P +
Sbjct: 256 SLFFPPLPRKKGGGGRRRRKERKKRKQKTPPPPPPPPRPFIY 381
Score = 29.6 bits (65), Expect = 4.3
Identities = 21/67 (31%), Positives = 27/67 (39%), Gaps = 10/67 (14%)
Frame = +1
Query: 421 PMPLFVQQCALEKRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPP----------PPP 470
P PL + E++ P PP KTK++ + PPPPP PP
Sbjct: 28 PPPLPQSRGRGEEKTQTHPPPP---------KTKKKKTTPPPPPPPP*GGGGNYAPPAPP 180
Query: 471 PPPMSFA 477
PPP A
Sbjct: 181 PPPSHMA 201
>BQ141909 weakly similar to GP|14253159|emb VMP3 protein {Volvox carteri f.
nagariensis}, partial (13%)
Length = 1223
Score = 38.9 bits (89), Expect = 0.007
Identities = 22/55 (40%), Positives = 27/55 (49%)
Frame = -1
Query: 421 PMPLFVQQCALEKRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMS 475
P LF+ + + +P+PPP PS S T P PPPPPPPPP S
Sbjct: 320 PFLLFLPPLSSPSPSSPLPSPPP-PSSLFPSPTPP-------PLPPPPPPPPPAS 180
Score = 32.3 bits (72), Expect = 0.67
Identities = 15/32 (46%), Positives = 17/32 (52%)
Frame = -2
Query: 442 PPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P R S S++ S PPPPPPP PPP
Sbjct: 352 PLRSSPSLTLPPSFCSSPPFLPPPPPPPSPPP 257
Score = 32.3 bits (72), Expect = 0.67
Identities = 23/76 (30%), Positives = 30/76 (39%), Gaps = 4/76 (5%)
Frame = -2
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFASRGNTA----MVKRAPQVVEL 494
P PPP PS + + PP PP PP PP + +S T+ R V
Sbjct: 262 PPPPPPPSSPLRPPLRSP------PPLPPLPPLPPFTPSSHLTTSSPPPSFLRCISSVVF 101
Query: 495 YHSLMKRDSRRDSSSG 510
L KRD ++ G
Sbjct: 100 IFPLRKRDGKKKKIGG 53
>BQ141925 homologue to GP|10727150|gb| Rm62 gene product {alt 1} [Drosophila
melanogaster], partial (3%)
Length = 1059
Score = 38.9 bits (89), Expect = 0.007
Identities = 14/17 (82%), Positives = 14/17 (82%)
Frame = +3
Query: 463 PPPPPPPPPPPMSFASR 479
PPPPPPPPPPP SF R
Sbjct: 21 PPPPPPPPPPPFSFPIR 71
Score = 35.8 bits (81), Expect = 0.060
Identities = 12/15 (80%), Positives = 13/15 (86%)
Frame = +3
Query: 463 PPPPPPPPPPPMSFA 477
PPPPPPPPPPP F+
Sbjct: 15 PPPPPPPPPPPPPFS 59
Score = 34.3 bits (77), Expect = 0.18
Identities = 11/11 (100%), Positives = 11/11 (100%)
Frame = +3
Query: 463 PPPPPPPPPPP 473
PPPPPPPPPPP
Sbjct: 12 PPPPPPPPPPP 44
Score = 32.3 bits (72), Expect = 0.67
Identities = 14/33 (42%), Positives = 17/33 (51%)
Frame = +2
Query: 463 PPPPPPPPPPPMSFASRGNTAMVKRAPQVVELY 495
PP PPPPPPPP S + Q +EL+
Sbjct: 17 PPAPPPPPPPPSFLFSNPFSKFPSLREQKLELH 115
Score = 31.6 bits (70), Expect = 1.1
Identities = 11/13 (84%), Positives = 11/13 (84%)
Frame = +1
Query: 461 VQPPPPPPPPPPP 473
V PPP PPPPPPP
Sbjct: 7 VLPPPRPPPPPPP 45
>BQ144482 similar to GP|6523547|emb| hydroxyproline-rich glycoprotein DZ-HRGP
{Volvox carteri f. nagariensis}, partial (16%)
Length = 993
Score = 38.5 bits (88), Expect = 0.009
Identities = 18/47 (38%), Positives = 21/47 (44%), Gaps = 8/47 (17%)
Frame = +1
Query: 439 PNPPPRPSCSISSKTKQECSAQVQP--------PPPPPPPPPPMSFA 477
P PP P + SA P PPPPPPPPPP+S +
Sbjct: 157 PRPPKSPRAHLFPPPSPSISAPPPPLPPLPLFRPPPPPPPPPPLSLS 297
Score = 37.0 bits (84), Expect = 0.027
Identities = 17/37 (45%), Positives = 18/37 (47%)
Frame = +1
Query: 441 PPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFA 477
PPP PS S PPPPPPPPP +S A
Sbjct: 193 PPPSPSISAPPPPLPPLPLFRPPPPPPPPPPLSLSLA 303
Score = 34.3 bits (77), Expect = 0.18
Identities = 18/48 (37%), Positives = 21/48 (43%)
Frame = +3
Query: 433 KRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFASRG 480
K A R P PPP P + + + PP P PPPPPP G
Sbjct: 168 KVAPRPPFPPPLPFY-LRPPSPSPSPPPLSPPSPSPPPPPPQPLLGPG 308
Score = 34.3 bits (77), Expect = 0.18
Identities = 20/51 (39%), Positives = 22/51 (42%)
Frame = +1
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFASRGNTAMVKRAP 489
P PPP P S+S A P P PP PPPP +R A V P
Sbjct: 262 PPPPPPPPLSLSL-----APAFAPPAPGPPRPPPPPRPPARPRPARVLAGP 399
Score = 33.5 bits (75), Expect = 0.30
Identities = 19/53 (35%), Positives = 23/53 (42%)
Frame = +1
Query: 421 PMPLFVQQCALEKRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P+PLF R P PPP S S++ +PPPPP PP P
Sbjct: 238 PLPLF--------RPPPPPPPPPPLSLSLAPAFAPPAPGPPRPPPPPRPPARP 372
Score = 32.3 bits (72), Expect = 0.67
Identities = 17/50 (34%), Positives = 22/50 (44%)
Frame = +3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFASRGNTAMVKRA 488
P PPP+P + +PP PPPP P P SR + +RA
Sbjct: 273 PPPPPQPLLGPGLRPPGP-----RPPAPPPPAPAPRPAPSRPSVGRPRRA 407
Score = 30.4 bits (67), Expect = 2.5
Identities = 18/56 (32%), Positives = 23/56 (40%)
Frame = +2
Query: 419 SVPMPLFVQQCALEKRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPM 474
S+P P F L +P PPP + + A PPP P PPP P+
Sbjct: 233 SLPSPSFAP--------LPLPPPPPPSASPWPRPSPPRPPAPRAPPPRPGPPPGPV 376
>BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (39%)
Length = 1358
Score = 38.5 bits (88), Expect = 0.009
Identities = 23/69 (33%), Positives = 29/69 (41%)
Frame = -2
Query: 421 PMPLFVQQCALEKRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFASRG 480
P+PL A R P PPP P +S + PP PPPPPPP +
Sbjct: 250 PLPL-----ASRLRPRASPPPPPPPPGPAASAPPRPPPPPPPPPQRPPPPPPPHTHQPEP 86
Query: 481 NTAMVKRAP 489
T+ +R P
Sbjct: 85 PTSPPRRQP 59
Score = 37.4 bits (85), Expect = 0.021
Identities = 21/69 (30%), Positives = 31/69 (44%)
Frame = -2
Query: 419 SVPMPLFVQQCALEKRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFAS 478
S P+ CA + P PPP+ + +T A+ PPPP PPP P+
Sbjct: 829 STPLDRARACCARRPPSPHRPAPPPQAHPARERRTAS--LARTPPPPPSSPPPAPLLPPR 656
Query: 479 RGNTAMVKR 487
GN +++ R
Sbjct: 655 NGNASLLPR 629
Score = 36.2 bits (82), Expect = 0.046
Identities = 16/37 (43%), Positives = 18/37 (48%), Gaps = 2/37 (5%)
Frame = -3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPP--PPPPP 473
P PPP P S + + PPPPPP PPPP
Sbjct: 159 PRPPPHPPRSGPPRPHPPTRTSLSPPPPPPAASPPPP 49
Score = 36.2 bits (82), Expect = 0.046
Identities = 15/36 (41%), Positives = 20/36 (54%)
Frame = -3
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPM 474
P P PRP + + + +PPPP PPPPPP+
Sbjct: 270 PPPRPRPHSP*QAACGRGHPPRHRPPPPGPPPPPPL 163
Score = 34.3 bits (77), Expect = 0.18
Identities = 19/46 (41%), Positives = 23/46 (49%), Gaps = 9/46 (19%)
Frame = -2
Query: 437 RIPNPPPRPSCSISSKTKQECSAQVQPPPPPP---------PPPPP 473
R P PPP P ++S+ + S PPPPPP PPPPP
Sbjct: 274 RAPPPPPAP-LPLASRLRPRASPP--PPPPPPGPAASAPPRPPPPP 146
Score = 32.0 bits (71), Expect = 0.87
Identities = 19/67 (28%), Positives = 24/67 (35%), Gaps = 16/67 (23%)
Frame = -3
Query: 439 PNPPPRPSCSISSKTKQEC----------------SAQVQPPPPPPPPPPPMSFASRGNT 482
P PPP PS T+ C S + PP PPP PP + +
Sbjct: 690 PPPPPPPSSPPGMGTRPSCQENWWAAPPGARPSRPSRYIPPPTSPPPASPPAPVPPQSPS 511
Query: 483 AMVKRAP 489
+ RAP
Sbjct: 510 TIRGRAP 490
Score = 30.8 bits (68), Expect = 1.9
Identities = 13/21 (61%), Positives = 14/21 (65%), Gaps = 4/21 (19%)
Frame = -2
Query: 463 PPPP----PPPPPPPMSFASR 479
PPPP PPPPP P+ ASR
Sbjct: 292 PPPPLDRAPPPPPAPLPLASR 230
Score = 30.4 bits (67), Expect = 2.5
Identities = 17/42 (40%), Positives = 18/42 (42%), Gaps = 3/42 (7%)
Frame = -3
Query: 441 PPPRPSCSISSKTKQECSAQVQPP---PPPPPPPPPMSFASR 479
PPPRP Q + PP PPPP PPPP R
Sbjct: 270 PPPRPR---PHSP*QAACGRGHPPRHRPPPPGPPPPPPLGPR 154
Score = 30.0 bits (66), Expect = 3.3
Identities = 15/39 (38%), Positives = 18/39 (45%)
Frame = -2
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFA 477
P PPPRP + A+ P PP PP PP + A
Sbjct: 499 PRPPPRPPPA--------APARAPPSGPPAPPTPPATRA 407
Score = 29.6 bits (65), Expect = 4.3
Identities = 16/48 (33%), Positives = 22/48 (45%)
Frame = -2
Query: 442 PPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFASRGNTAMVKRAP 489
PP P+ + + + C A+ P P P PPP A TA + R P
Sbjct: 844 PPDPASTPLDRARA-CCARRPPSPHRPAPPPQAHPARERRTASLARTP 704
Score = 29.6 bits (65), Expect = 4.3
Identities = 18/50 (36%), Positives = 23/50 (46%), Gaps = 7/50 (14%)
Frame = -1
Query: 434 RALRIPN----PPPRPSCSISSKTKQECSA---QVQPPPPPPPPPPPMSF 476
R LR P+ PP P+ + + T A + PPPP P PP P F
Sbjct: 182 RRLRPPSAPAPPPTPPAAAPPAPTPPHAPA*APHLPPPPPAPRPPRPPPF 33
Score = 29.3 bits (64), Expect = 5.7
Identities = 17/58 (29%), Positives = 25/58 (42%)
Frame = -1
Query: 421 PMPLFVQQCALEKRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFAS 478
P P ++ L K+ P P P +++ + PPPP PP PP S A+
Sbjct: 668 PPPPEWERVPLAKKIGGQPRPALVPLVRVATYPRPPRPPPPVPPPPSPPRAPPQSEAA 495
Score = 28.9 bits (63), Expect = 7.4
Identities = 20/58 (34%), Positives = 26/58 (44%), Gaps = 4/58 (6%)
Frame = -1
Query: 437 RIPNPPPRPSCSISSKTKQECSAQVQPPPPPP----PPPPPMSFASRGNTAMVKRAPQ 490
R P PPP P S + ++ PPPPPP P P+ S + A RAP+
Sbjct: 569 RPPRPPP-PVPPPPSPPRAPPQSEAAPPPPPPPRGARPRAPLRPPSAPDPAGHPRAPR 399
Score = 28.9 bits (63), Expect = 7.4
Identities = 13/38 (34%), Positives = 15/38 (39%)
Frame = -1
Query: 434 RALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPP 471
R P PP P + Q + PPPPPPP
Sbjct: 587 RVATYPRPPRPPPPVPPPPSPPRAPPQSEAAPPPPPPP 474
Score = 28.5 bits (62), Expect = 9.6
Identities = 17/68 (25%), Positives = 24/68 (35%), Gaps = 18/68 (26%)
Frame = -3
Query: 441 PPPRPSCSISSKTKQECSAQVQPPPPPP------------------PPPPPMSFASRGNT 482
PP P + ++ + PP PPP PP P S RG
Sbjct: 555 PPASPPAPVPPQSPSTIRGRAPPPAPPPRRPPARPPPAPQRPRPRRPPARPPSGRRRGAR 376
Query: 483 AMVKRAPQ 490
A V++ P+
Sbjct: 375 AKVRKLPK 352
>TC87201 similar to PIR|F86388|F86388 hypothetical protein AAG29221.1
[imported] - Arabidopsis thaliana, partial (41%)
Length = 991
Score = 38.5 bits (88), Expect = 0.009
Identities = 18/49 (36%), Positives = 22/49 (44%)
Frame = +1
Query: 441 PPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFASRGNTAMVKRAP 489
PPP P I + V P PPPP PPPP + S V ++P
Sbjct: 55 PPPSPPPPIRIQITTRAPPYVYPSPPPPSPPPPYIYKSPPPPPYVYKSP 201
Score = 32.7 bits (73), Expect = 0.51
Identities = 14/33 (42%), Positives = 15/33 (45%)
Frame = +1
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPP 471
P P P P I T+ PPPP PPPP
Sbjct: 55 PPPSPPPPIRIQITTRAPPYVYPSPPPPSPPPP 153
Score = 31.2 bits (69), Expect = 1.5
Identities = 16/39 (41%), Positives = 18/39 (46%)
Frame = +1
Query: 440 NPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFAS 478
+PPP P S V P PPPP PPPP + S
Sbjct: 166 SPPPPPYVYKSPPPPP----YVYPSPPPPSPPPPYIYKS 270
Score = 30.4 bits (67), Expect = 2.5
Identities = 12/27 (44%), Positives = 15/27 (55%)
Frame = +1
Query: 463 PPPPPPPPPPPMSFASRGNTAMVKRAP 489
PPPP P PPPP + S V ++P
Sbjct: 322 PPPPSPSPPPPYVYKSPPPPPYVYKSP 402
Score = 29.6 bits (65), Expect = 4.3
Identities = 10/14 (71%), Positives = 11/14 (78%)
Frame = +1
Query: 461 VQPPPPPPPPPPPM 474
V P PPPP PPPP+
Sbjct: 40 VYPSPPPPSPPPPI 81
Score = 28.9 bits (63), Expect = 7.4
Identities = 19/56 (33%), Positives = 22/56 (38%)
Frame = +1
Query: 418 ESVPMPLFVQQCALEKRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
ES P P +V + +PPP P S PPPP P PPPP
Sbjct: 424 ESPPPPQYVYK-----------SPPPPPYVYES------------PPPPSPSPPPP 522
>BQ144407 weakly similar to GP|6523547|emb| hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (23%)
Length = 1217
Score = 38.1 bits (87), Expect = 0.012
Identities = 21/57 (36%), Positives = 28/57 (48%), Gaps = 2/57 (3%)
Frame = -1
Query: 419 SVPMPLFVQQCALEKRALRIPNPPPRPSCSISSKTKQECSAQ--VQPPPPPPPPPPP 473
++P+P + I PPPR S S++K S + PPPP PPPPPP
Sbjct: 356 AIPLPPRFMPLIGSPTSSSIRIPPPRSSPYRCSRSKAPISPPFLLPPPPPRPPPPPP 186
Score = 35.0 bits (79), Expect = 0.10
Identities = 19/56 (33%), Positives = 24/56 (41%)
Frame = -2
Query: 435 ALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFASRGNTAMVKRAPQ 490
AL P PPPRP+ C P PPP PP +F RG+ + P+
Sbjct: 412 ALSSPTPPPRPAAHSLLPPPFRC-PPASCPLSAPPPLPPSAFPPRGHPPIAAPGPR 248
Score = 28.5 bits (62), Expect = 9.6
Identities = 14/36 (38%), Positives = 15/36 (40%)
Frame = -1
Query: 438 IPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
+P PPPRP PPPPP P PP
Sbjct: 224 LPPPPPRP-----------------PPPPPGRPRPP 168
Score = 28.5 bits (62), Expect = 9.6
Identities = 13/36 (36%), Positives = 19/36 (52%), Gaps = 1/36 (2%)
Frame = -3
Query: 463 PPPPPPPPPPPMSFASRGNTAM-VKRAPQVVELYHS 497
PP PPPPP PP A + ++ + AP + + S
Sbjct: 219 PPTPPPPPAPPRPAAPPPSLSISINPAPHTIYILPS 112
Score = 28.5 bits (62), Expect = 9.6
Identities = 14/36 (38%), Positives = 20/36 (54%)
Frame = -3
Query: 440 NPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMS 475
+PPP SI++ + + S + P P PPPP P S
Sbjct: 636 SPPPSLDISITAPSTRPSS--LTPTPASPPPPIPRS 535
>TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana, partial (91%)
Length = 860
Score = 38.1 bits (87), Expect = 0.012
Identities = 29/96 (30%), Positives = 36/96 (37%), Gaps = 20/96 (20%)
Frame = -2
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPM--------------------SFAS 478
P PPPRP PPPPPPPPPP+ S AS
Sbjct: 316 PPPPPRPR-----------------PPPPPPPPPPLLEL*ESSTLHPFLPSRSRIASCAS 188
Query: 479 RGNTAMVKRAPQVVELYHSLMKRDSRRDSSSGGLSD 514
++ +K P + +L R+SSS LSD
Sbjct: 187 LRSSKSIKA*PGGLRATQTLRITPKTRNSSSRSLSD 80
>TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear protein.
[strain B95-8 Human herpesvirus 4] {Epstein-barr
virus}, partial (23%)
Length = 431
Score = 38.1 bits (87), Expect = 0.012
Identities = 19/44 (43%), Positives = 21/44 (47%)
Frame = -1
Query: 430 ALEKRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
AL + P PPP P I C+ PPPP PPPPPP
Sbjct: 269 ALNPQGSYPPWPPPSPP-PIPPPYAPPCAPPPPPPPPEPPPPPP 141
Score = 35.8 bits (81), Expect = 0.060
Identities = 16/35 (45%), Positives = 18/35 (50%)
Frame = -1
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P + C+ PPPPPP PPPP
Sbjct: 110 PVPPPLPYPT-------PCAPPAPPPPPPPLPPPP 27
Score = 33.9 bits (76), Expect = 0.23
Identities = 16/37 (43%), Positives = 18/37 (48%), Gaps = 2/37 (5%)
Frame = -1
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPP--PPP 473
P+PPP P +PPPPPPPP PPP
Sbjct: 230 PSPPPIPPPYAPPCAPPPPPPPPEPPPPPPPPGDPPP 120
Score = 30.4 bits (67), Expect = 2.5
Identities = 14/38 (36%), Positives = 16/38 (41%)
Frame = -1
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSF 476
P PPP P PPP PPP PPP+ +
Sbjct: 155 PPPPPPPG---------------DPPPYPPPVPPPLPY 87
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.312 0.128 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,912,591
Number of Sequences: 36976
Number of extensions: 416333
Number of successful extensions: 14938
Number of sequences better than 10.0: 464
Number of HSP's better than 10.0 without gapping: 4209
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8060
length of query: 732
length of database: 9,014,727
effective HSP length: 103
effective length of query: 629
effective length of database: 5,206,199
effective search space: 3274699171
effective search space used: 3274699171
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC124968.5


