

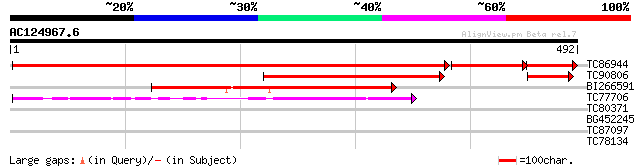
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124967.6 - phase: 0 /pseudo
(492 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86944 similar to GP|15215806|gb|AAK91448.1 At3g14201 {Arabidop... 776 0.0
TC90806 similar to GP|15217295|gb|AAK92639.1 Putative phosphoino... 230 1e-60
BI266591 similar to GP|19424035|gb unknown protein {Arabidopsis ... 192 3e-49
TC77706 similar to GP|14532908|gb|AAK64136.1 unknown protein {Ar... 103 2e-22
TC80371 similar to GP|14326543|gb|AAK60316.1 AT4g18020/T6K21_200... 31 1.2
BG452245 similar to PIR|H86341|H8 hypothetical protein AAD30599.... 30 2.8
TC87097 similar to GP|14334540|gb|AAK59678.1 putative protein {A... 29 4.7
TC78134 similar to GP|9795609|gb|AAF98427.1| Unknown protein {Ar... 28 8.0
>TC86944 similar to GP|15215806|gb|AAK91448.1 At3g14201 {Arabidopsis
thaliana}, partial (50%)
Length = 2552
Score = 776 bits (2005), Expect(3) = 0.0
Identities = 378/379 (99%), Positives = 379/379 (99%)
Frame = +2
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 62
RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP
Sbjct: 290 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 469
Query: 63 DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEFANAIDFINK 122
DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEFANAIDFINK
Sbjct: 470 DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEFANAIDFINK 649
Query: 123 DLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTLRPEDCLKW 182
DLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTLRPEDCLKW
Sbjct: 650 DLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTLRPEDCLKW 829
Query: 183 PSTDDVDKGSFSPTVHVEYDNEDANDLERKPSDEINVSNENHSAKPPRLQNGVLRTNCID 242
PSTDDVDKGSFSPTVHVEYDNEDANDL+RKPSDEINVSNENHSAKPPRLQNGVLRTNCID
Sbjct: 830 PSTDDVDKGSFSPTVHVEYDNEDANDLQRKPSDEINVSNENHSAKPPRLQNGVLRTNCID 1009
Query: 243 CLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQFYERMGDTLAHQYGGS 302
CLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQFYERMGDTLAHQYGGS
Sbjct: 1010CLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQFYERMGDTLAHQYGGS 1189
Query: 303 AAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINVFLGHFQPQQDKPAL 362
AAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINVFLGHFQPQQDKPAL
Sbjct: 1190AAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINVFLGHFQPQQDKPAL 1369
Query: 363 WELGSDQHYDTGRIGDDDA 381
WELGSDQHYDTGRIGDDDA
Sbjct: 1370WELGSDQHYDTGRIGDDDA 1426
Score = 130 bits (328), Expect(3) = 0.0
Identities = 66/66 (100%), Positives = 66/66 (100%)
Frame = +1
Query: 384 VIFQKIFLRWKHSS*QFYTYASFKCQGRKILKPRFTRPIRRREQGLL*VLT*NIYY*N*E 443
VIFQKIFLRWKHSS*QFYTYASFKCQGRKILKPRFTRPIRRREQGLL*VLT*NIYY*N*E
Sbjct: 1429 VIFQKIFLRWKHSS*QFYTYASFKCQGRKILKPRFTRPIRRREQGLL*VLT*NIYY*N*E 1608
Query: 444 *HFIFK 449
*HFIFK
Sbjct: 1609 *HFIFK 1626
Score = 99.0 bits (245), Expect(3) = 0.0
Identities = 43/44 (97%), Positives = 44/44 (99%)
Frame = +2
Query: 449 KKERCAESHHIYYSEHGDSFSCSNFVDLDWLSSSANSCEEDPYE 492
+KERCAESHHIYYSEHGDSFSCSNFVDLDWLSSSANSCEEDPYE
Sbjct: 1670 QKERCAESHHIYYSEHGDSFSCSNFVDLDWLSSSANSCEEDPYE 1801
>TC90806 similar to GP|15217295|gb|AAK92639.1 Putative phosphoinositide
phosphatase {Oryza sativa} [Oryza sativa (japonica
cultivar-group)], partial (21%)
Length = 1285
Score = 230 bits (586), Expect = 1e-60
Identities = 109/157 (69%), Positives = 125/157 (79%)
Frame = +1
Query: 221 NENHSAKPPRLQNGVLRTNCIDCLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPV 280
N++++ K +Q+GVLRTNCIDCLDRTNVAQYAYGL A+G QLH+LG IE P I L +P+
Sbjct: 256 NKDYNVKAKTVQSGVLRTNCIDCLDRTNVAQYAYGLVALGRQLHALGFIESPHIKLGNPL 435
Query: 281 ANDLMQFYERMGDTLAHQYGGSAAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDA 340
A DLM+ YE MGDTLA QYGGSAAHNKIFS RGQW+AA QSQE RTLQRYY+N + D
Sbjct: 436 AKDLMKVYESMGDTLAFQYGGSAAHNKIFSRSRGQWKAAAQSQELIRTLQRYYNNTYTDG 615
Query: 341 EKQDAINVFLGHFQPQQDKPALWELGSDQHYDTGRIG 377
KQ AIN+FLGHFQPQQ KP LW+L SDQHY G G
Sbjct: 616 FKQKAINIFLGHFQPQQGKPVLWKLDSDQHYLLGSHG 726
Score = 58.2 bits (139), Expect = 7e-09
Identities = 24/40 (60%), Positives = 31/40 (77%)
Frame = +2
Query: 450 KERCAESHHIYYSEHGDSFSCSNFVDLDWLSSSANSCEED 489
K++ E+ HI Y EH D+ CSNF+D+DWLSSS NSCEE+
Sbjct: 947 KDQYCENDHICYDEHEDANDCSNFLDVDWLSSSENSCEEE 1066
>BI266591 similar to GP|19424035|gb unknown protein {Arabidopsis thaliana},
partial (20%)
Length = 666
Score = 192 bits (487), Expect = 3e-49
Identities = 100/221 (45%), Positives = 142/221 (64%), Gaps = 9/221 (4%)
Frame = +1
Query: 124 LSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTLRPEDCLKWP 183
L E+ LRF+HWD HK ++K+ NVL +LG VA+ AL LT F+Y + ++ +
Sbjct: 7 LPAESHLRFIHWDFHKFAKTKSANVLAVLGAVASEALDLTSFYYSGKPNIVKRANKSNRT 186
Query: 184 STD------DVDKGSFSPTVHVEYDNEDANDLERKPSD-EINVSNENH--SAKPPRLQNG 234
ST D+ S + +E N + + + ++N N++ ++ P Q+G
Sbjct: 187 STARDASLRDLTASS-GDLARLGISSEVLNSMANRDRETDMNHQNKDDYFNSDTPHFQSG 363
Query: 235 VLRTNCIDCLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQFYERMGDT 294
VLRTNCIDCLDRTNVAQYAYGL A+G QLH++G+ + PK+D D +A LM Y+ MGD
Sbjct: 364 VLRTNCIDCLDRTNVAQYAYGLQALGRQLHAMGLSDVPKVDPDSSIAAALMDMYQSMGDA 543
Query: 295 LAHQYGGSAAHNKIFSARRGQWRAATQSQEFFRTLQRYYSN 335
LA QYGGSAAHN +F R+G+W+A TQS+EF ++++RYYSN
Sbjct: 544 LAQQYGGSAAHNTVFPERQGKWKATTQSREFLKSIKRYYSN 666
>TC77706 similar to GP|14532908|gb|AAK64136.1 unknown protein {Arabidopsis
thaliana}, partial (98%)
Length = 2358
Score = 103 bits (256), Expect = 2e-22
Identities = 101/353 (28%), Positives = 151/353 (42%), Gaps = 2/353 (0%)
Frame = +1
Query: 3 RYLRRGVNEKGRVANDVETEQIV-FEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIK 61
R RRG + G VAN VETEQ++ F +S +Q RGSIPL W Q+ L K
Sbjct: 859 RMWRRGADPDGYVANFVETEQLMQFNGY-------TASFVQIRGSIPLLW-QQIVDLTYK 1014
Query: 62 PDIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEFANAIDFIN 121
P L K +++ + + H +L K+YG ++ ++L+ H + R L ++F + + +
Sbjct: 1015 PKFELLKLEEAPRVLERHILDLRKKYG-AVLAVDLVNKHGGEGR---LCEKFGSTMQHVA 1182
Query: 122 KDLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTLRPEDCLK 181
D +R++H+D H + G V L++ Y Q+S
Sbjct: 1183 SD-----DVRYVHFDFHH-----------ICGHVHFERLSM---LYDQIS---------- 1275
Query: 182 WPSTDDVDKGSFSPTVHVEYDNEDANDLERKPSDEINVSNENHSAKPPRLQNGVLRTNCI 241
+ LER +N E + Q GV+RTNCI
Sbjct: 1276 -------------------------DFLERNGYLLLNEKGEK-----MKEQLGVVRTNCI 1365
Query: 242 DCLDRTNVAQYAYGLAAIGHQLHSLGII-EHPKIDLDDPVANDLMQFYERMGDTLAHQYG 300
DCLDRTNV Q G + +QL LG+ I + + GD ++ QY
Sbjct: 1366 DCLDRTNVTQSMIGRNMLEYQLRRLGVFGAEETISSHSNLDERFKILWANHGDDISIQYS 1545
Query: 301 GSAAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINVFLGHF 353
G+ A F R G + LQRYY N ++D KQDAI++ GH+
Sbjct: 1546 GTPALKGDF-VRFGHRTVQGIINDGCNALQRYYLNNFVDGTKQDAIDLLQGHY 1701
>TC80371 similar to GP|14326543|gb|AAK60316.1 AT4g18020/T6K21_200
{Arabidopsis thaliana}, partial (39%)
Length = 1352
Score = 30.8 bits (68), Expect = 1.2
Identities = 31/133 (23%), Positives = 54/133 (40%)
Frame = +3
Query: 116 AIDFINKDLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTLR 175
A++F+ K LSE+ +HK F ++A+ + L V ++ + Q +TL
Sbjct: 381 AVEFLTKPLSEDKLKNIWQHVVHKAFNAEASALSESLKPVKE---SVESMLHLQTDNTLH 551
Query: 176 PEDCLKWPSTDDVDKGSFSPTVHVEYDNEDANDLERKPSDEINVSNENHSAKPPRLQNGV 235
ST +D S E+++ A+D PS P+L+ G
Sbjct: 552 -------ESTISIDLDKVSKFSDNEHEHSAASDKYPAPS-------------TPQLKQGA 671
Query: 236 LRTNCIDCLDRTN 248
+ DC ++TN
Sbjct: 672 RLVDDGDCHEQTN 710
>BG452245 similar to PIR|H86341|H8 hypothetical protein AAD30599.1 [imported]
- Arabidopsis thaliana, partial (18%)
Length = 653
Score = 29.6 bits (65), Expect = 2.8
Identities = 15/49 (30%), Positives = 25/49 (50%)
Frame = +1
Query: 266 LGIIEHPKIDLDDPVANDLMQFYERMGDTLAHQYGGSAAHNKIFSARRG 314
+G+I P +L + +D+ +F + MG YGGS +I +RG
Sbjct: 244 IGLIMAPTRELVQQIHSDIRKFTKVMGIRCVPVYGGSGVAQQISELKRG 390
>TC87097 similar to GP|14334540|gb|AAK59678.1 putative protein {Arabidopsis
thaliana}, partial (62%)
Length = 1234
Score = 28.9 bits (63), Expect = 4.7
Identities = 25/74 (33%), Positives = 32/74 (42%), Gaps = 6/74 (8%)
Frame = +2
Query: 87 YGNPIIILNLIKTHEKKPREAILRQEFANAIDFINKDLSEENRLRF------LHWDLHKH 140
YGN I ILNL H + LR E + + E RLR LH L+K
Sbjct: 116 YGNAIAILNLPHLHLPPKKLIRLRSENLLHVPLLLAACVGE-RLRLVLALLQLHLQLNKS 292
Query: 141 FQSKATNVLLLLGK 154
+ K T LL++ K
Sbjct: 293 LEMKVTTKLLVMSK 334
>TC78134 similar to GP|9795609|gb|AAF98427.1| Unknown protein {Arabidopsis
thaliana}, partial (46%)
Length = 1133
Score = 28.1 bits (61), Expect = 8.0
Identities = 13/31 (41%), Positives = 19/31 (60%)
Frame = +2
Query: 90 PIIILNLIKTHEKKPREAILRQEFANAIDFI 120
P+ +LN + EKK RE L ++ +A DFI
Sbjct: 47 PLSLLNFVFEREKKEREKSLPHQWKSAQDFI 139
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.328 0.141 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,617,442
Number of Sequences: 36976
Number of extensions: 244339
Number of successful extensions: 1291
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 1276
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1288
length of query: 492
length of database: 9,014,727
effective HSP length: 100
effective length of query: 392
effective length of database: 5,317,127
effective search space: 2084313784
effective search space used: 2084313784
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC124967.6


