

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
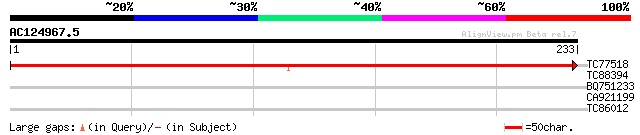
Query= AC124967.5 + phase: 0 /pseudo
(233 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77518 similar to SP|Q94HF1|IF3B_ORYSA Eukaryotic translation i... 414 e-116
TC88394 similar to PIR|T47576|T47576 FKBP12 interacting protein ... 28 2.3
BQ751233 similar to GP|18307448|em probable low-affinity hexose ... 27 5.1
CA921199 similar to PIR|T51950|T51 DNA-directed RNA polymerase (... 27 6.7
TC86012 homologue to PIR|T00795|T00795 26S proteasome regulatory... 27 6.7
>TC77518 similar to SP|Q94HF1|IF3B_ORYSA Eukaryotic translation initiation
factor 3 subunit 11 (eIF-3 p25) (eIF3k). [Rice] {Oryza
sativa}, partial (94%)
Length = 1020
Score = 414 bits (1065), Expect = e-116
Identities = 217/234 (92%), Positives = 219/234 (92%), Gaps = 1/234 (0%)
Frame = +1
Query: 1 MGREAAAAAEKGQGGTAYTVEQLVAFNRYNPDILPDLENYVNDQVSSQTYSLESNLCLLR 60
MGREAAAAAEKGQGGTAYTVEQLVAFNRYNPDILPDLENYVNDQVSSQTYSLESNLCLLR
Sbjct: 106 MGREAAAAAEKGQGGTAYTVEQLVAFNRYNPDILPDLENYVNDQVSSQTYSLESNLCLLR 285
Query: 61 LYQFESEKMSSQIVARILVKALMAMPAPDFSLCLFLIPERVIANGRSSSRH*L-FCLTI* 119
LYQFESEKMSSQIVARILVKALMAMPAPDFSLCLFLIPERV + + L L
Sbjct: 286 LYQFESEKMSSQIVARILVKALMAMPAPDFSLCLFLIPERVQMEEQFKTLIVLSHYLETG 465
Query: 120 RFRQFWDEASKSRHIVEAVPGFEQAIQGYAIHVLSLTYQKIPRTVLAEAINVEGLSLDKF 179
RFRQFWDEASKSRHIVEAVPGFEQAIQGYAIHVLSLTYQKIPRTVLAEAINVEGLSLDKF
Sbjct: 466 RFRQFWDEASKSRHIVEAVPGFEQAIQGYAIHVLSLTYQKIPRTVLAEAINVEGLSLDKF 645
Query: 180 LEHQVENSGWVIEKSQGKGQLIVLPRNEFNDPILKKNTADSVPLEHVTRIFPIL 233
LEHQVENSGWVIEKSQGKGQLIVLPRNEFNDPILKKNTADSVPLEHVTRIFPIL
Sbjct: 646 LEHQVENSGWVIEKSQGKGQLIVLPRNEFNDPILKKNTADSVPLEHVTRIFPIL 807
>TC88394 similar to PIR|T47576|T47576 FKBP12 interacting protein (FIP37) -
Arabidopsis thaliana, partial (56%)
Length = 1285
Score = 28.5 bits (62), Expect = 2.3
Identities = 14/42 (33%), Positives = 24/42 (56%)
Frame = -2
Query: 42 NDQVSSQTYSLESNLCLLRLYQFESEKMSSQIVARILVKALM 83
N+ S++T+ L S LC+LRL Q + + + R L+ L+
Sbjct: 1002 NEFHSAKTFKLTSELCILRLLQCKFHRQFMYLSFRSLISDLL 877
>BQ751233 similar to GP|18307448|em probable low-affinity hexose transporter
HXT3 {Neurospora crassa}, partial (30%)
Length = 659
Score = 27.3 bits (59), Expect = 5.1
Identities = 18/46 (39%), Positives = 23/46 (49%), Gaps = 7/46 (15%)
Frame = -1
Query: 164 VLAEAINV-------EGLSLDKFLEHQVENSGWVIEKSQGKGQLIV 202
VLAEA+ V L+ D EH+ N G EK GKG L++
Sbjct: 266 VLAEAVEVVLHEEESRDLTTD-ITEHEATNGGDETEKGSGKGDLLL 132
>CA921199 similar to PIR|T51950|T51 DNA-directed RNA polymerase (EC 2.7.7.6)
23K chain [imported] - Arabidopsis thaliana, partial
(49%)
Length = 756
Score = 26.9 bits (58), Expect = 6.7
Identities = 15/35 (42%), Positives = 18/35 (50%)
Frame = -3
Query: 41 VNDQVSSQTYSLESNLCLLRLYQFESEKMSSQIVA 75
VN VS+Q+YSL NLC +Q S VA
Sbjct: 493 VNAAVSNQSYSLCQNLCF*NCFQISLGSFSGGRVA 389
>TC86012 homologue to PIR|T00795|T00795 26S proteasome regulatory subunit
[imported] - Arabidopsis thaliana, partial (59%)
Length = 2134
Score = 26.9 bits (58), Expect = 6.7
Identities = 14/42 (33%), Positives = 24/42 (56%), Gaps = 1/42 (2%)
Frame = +3
Query: 57 CLLRLYQFESEKMSSQIVARILVKALMAMPAPDF-SLCLFLI 97
C+ + F + + Q V R+LVK +P+PD+ S+C L+
Sbjct: 702 CIHVSHSFVNLREYRQEVLRLLVKVFQKLPSPDYLSICQCLM 827
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.139 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,568,076
Number of Sequences: 36976
Number of extensions: 85088
Number of successful extensions: 402
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 401
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 401
length of query: 233
length of database: 9,014,727
effective HSP length: 93
effective length of query: 140
effective length of database: 5,575,959
effective search space: 780634260
effective search space used: 780634260
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC124967.5


