

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124967.11 + phase: 0
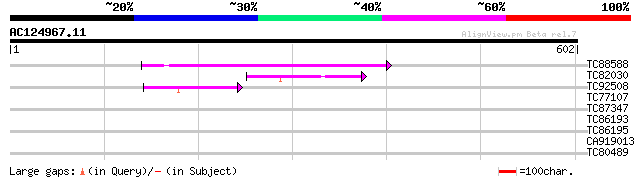
(602 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88588 similar to PIR|T08400|T08400 late embryonic abundant pro... 164 1e-40
TC82030 similar to GP|5091614|gb|AAD39602.1| Contains a PF|00561... 61 1e-09
TC92508 weakly similar to GP|5091614|gb|AAD39602.1| Contains a P... 52 9e-07
TC77107 ornithine aminotransferase [Medicago truncatula] 30 3.5
TC87347 similar to PIR|D86268|D86268 hypothetical protein AAF998... 29 4.5
TC86193 similar to GP|16974678|gb|AAL32439.1 SH3 domain-containi... 29 4.5
TC86195 similar to GP|16974678|gb|AAL32439.1 SH3 domain-containi... 29 4.5
CA919013 similar to PIR|C86475|C864 unknown protein 42479-41336... 29 5.9
TC80489 similar to GP|9294341|dbj|BAB02238.1 contains similarity... 28 7.7
>TC88588 similar to PIR|T08400|T08400 late embryonic abundant protein EMB8
homolog F18B3.70 - Arabidopsis thaliana, partial (83%)
Length = 1522
Score = 164 bits (414), Expect = 1e-40
Identities = 95/271 (35%), Positives = 154/271 (56%), Gaps = 6/271 (2%)
Frame = +1
Query: 141 ELKYQRVCLSASDGGVVSLDWPVELDLEEERGL--DSTLLIV-PGTPQGSMDDNIRVFVI 197
++ +R CL DGG V+LDW ++R L D+ LLI+ PG GS D +R +I
Sbjct: 247 DVTLRRHCLRTQDGGSVALDWVSG----DDRRLPPDAPLLILLPGLTGGSGDSYVRHMLI 414
Query: 198 DALKRGFFPVVMNPRGCASSPITTPRLFTAADSDDICTAITYINKARPWTTLMGVGWGYG 257
A +G+ VV N RGC SP+TTP+ ++A+ D+ +++++ P L GVGW G
Sbjct: 415 RARNKGWRVVVFNSRGCGGSPVTTPQFYSASFLGDMHEVVSHVSDRYPDANLYGVGWSLG 594
Query: 258 ANMLTKYLAEVGERTPLTAATCIDNPFDLDEATRTF--PYHHVTDQKLTRGLINILQTNK 315
AN+L +YL + PL+ A + NPFDL + F ++ + D+ L+ L I +
Sbjct: 595 ANILVRYLGQESHNCPLSGAVSLCNPFDLVVSDEDFRKGFNKIYDKALSSALSKIFNKHA 774
Query: 316 ALFQGKAKGXDVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDIKIPVLF 375
LF+ ++ AKSVRDF++ ++ VS+GF ++D+Y+ +S+ + IK ++ P+L
Sbjct: 775 LLFEDIGGEYNIPLVANAKSVRDFDDGLTRVSFGFKSVDDYYSNSSSSDSIKHVQTPLLC 954
Query: 376 IQSDNG-MVPAFSVPRNLIAENPFTSLLLCS 405
IQ+ N + P+ +PR I ENP L++ S
Sbjct: 955 IQAANDPIAPSRGIPREDIKENPNCLLVVTS 1047
>TC82030 similar to GP|5091614|gb|AAD39602.1| Contains a PF|00561 alpha/beta
hydrolase fold domain. {Arabidopsis thaliana}, partial
(37%)
Length = 1128
Score = 60.8 bits (146), Expect = 1e-09
Identities = 37/130 (28%), Positives = 65/130 (49%), Gaps = 3/130 (2%)
Frame = +1
Query: 252 VGWGYGANMLTKYLAEVGERTPLTAATCIDNPFDL---DEATRTFPYHHVTDQKLTRGLI 308
VG GAN+L KYL E GE TP+ A + +P+DL D P ++ L G
Sbjct: 13 VGTSIGANILVKYLGEDGENTPVAGAVAVCSPWDLLIADRFISRAPVQKFYNKALAGGQK 192
Query: 309 NILQTNKALFQGKAKGXDVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKD 368
+ + ++ F A + KAL SVRDF++ + + + ++ +Y + S+ ++
Sbjct: 193 DYAKLHQPQFTRLANWEGIEKAL---SVRDFDDHATRMVGKYETVDTYYRRCSSSTYVQS 363
Query: 369 IKIPVLFIQS 378
+ +P+L I +
Sbjct: 364 VSVPLLCISA 393
>TC92508 weakly similar to GP|5091614|gb|AAD39602.1| Contains a PF|00561
alpha/beta hydrolase fold domain. {Arabidopsis
thaliana}, partial (27%)
Length = 731
Score = 51.6 bits (122), Expect = 9e-07
Identities = 31/115 (26%), Positives = 50/115 (42%), Gaps = 10/115 (8%)
Frame = +2
Query: 143 KYQRVCLSASDGGVVSLDWPVELDLEEERGLDSTL----------LIVPGTPQGSMDDNI 192
KY+R + SDGG ++LDW D+ D + + +PG S +
Sbjct: 365 KYKRQLFTTSDGGTLALDWVTSSDVSGSVHHDDDVVTKDDSTPIVICIPGLTSDSSSPYL 544
Query: 193 RVFVIDALKRGFFPVVMNPRGCASSPITTPRLFTAADSDDICTAITYINKARPWT 247
+ KRG+ VV N RG PIT+ + + ++D T + Y++ P T
Sbjct: 545 KHLAYHTAKRGWKVVVSNHRGFGGVPITSDCFYNSGWTEDTRTVVNYLHNRIPST 709
>TC77107 ornithine aminotransferase [Medicago truncatula]
Length = 1681
Score = 29.6 bits (65), Expect = 3.5
Identities = 22/85 (25%), Positives = 34/85 (39%), Gaps = 5/85 (5%)
Frame = -3
Query: 24 YKRRRLKIKASFPVPPPSPF-----ENLFNTLISQCSSVNSIDFIAPSLGFASGSALFFS 78
Y+ + +K + P SP LF L+ Q + S G + +LFF
Sbjct: 1256 YRGQTFAVKLNSNEQPSSPHLLHMVRVLFLNLLQQTEKLTS------QFGRSLSKSLFFD 1095
Query: 79 RFKSSQNSDVGEWILFASPTPFNRF 103
FK + G+WI S + +RF
Sbjct: 1094 HFK*GNGNCTGQWISTKSTSMLSRF 1020
>TC87347 similar to PIR|D86268|D86268 hypothetical protein AAF99821.1
[imported] - Arabidopsis thaliana, partial (8%)
Length = 1007
Score = 29.3 bits (64), Expect = 4.5
Identities = 24/66 (36%), Positives = 31/66 (46%), Gaps = 8/66 (12%)
Frame = -2
Query: 33 ASFPVPPPSP---FENLFNTLISQCSSVNSIDFIA-----PSLGFASGSALFFSRFKSSQ 84
A+ PPSP FEN+F+ L S SS N FI PSL G F + S Q
Sbjct: 469 ATSTTTPPSPLFSFENIFSHLCSSVSS*NIDIFICSFKFIPSLTNIKGQYHFPTTTDSEQ 290
Query: 85 NSDVGE 90
+ +G+
Sbjct: 289 SFKIGK 272
>TC86193 similar to GP|16974678|gb|AAL32439.1 SH3 domain-containing protein 2
{Arabidopsis thaliana}, partial (97%)
Length = 1311
Score = 29.3 bits (64), Expect = 4.5
Identities = 20/64 (31%), Positives = 31/64 (48%), Gaps = 2/64 (3%)
Frame = +1
Query: 37 VPPPSPFENLFNTLISQCSSVNSIDFIAPSLGFASGSALF-FSRFKSSQ-NSDVGEWILF 94
+PPP P+E + SQ + S D S+G+ G LF +S + N VG++I+
Sbjct: 943 MPPPPPYEEVNGVYASQTTHNGSTD----SMGYFLGEVLFPYSAVSEVELNLSVGDYIVI 1110
Query: 95 ASPT 98
T
Sbjct: 1111 RKVT 1122
>TC86195 similar to GP|16974678|gb|AAL32439.1 SH3 domain-containing protein
2 {Arabidopsis thaliana}, partial (58%)
Length = 848
Score = 29.3 bits (64), Expect = 4.5
Identities = 20/64 (31%), Positives = 31/64 (48%), Gaps = 2/64 (3%)
Frame = +2
Query: 37 VPPPSPFENLFNTLISQCSSVNSIDFIAPSLGFASGSALF-FSRFKSSQ-NSDVGEWILF 94
+PPP P+E + SQ + S D S+G+ G LF +S + N VG++I+
Sbjct: 392 MPPPPPYEEVNGVYASQTTHNGSTD----SMGYFLGEVLFPYSAVSEVELNLSVGDYIVI 559
Query: 95 ASPT 98
T
Sbjct: 560 RKVT 571
>CA919013 similar to PIR|C86475|C864 unknown protein 42479-41336 [imported]
- Arabidopsis thaliana, partial (25%)
Length = 471
Score = 28.9 bits (63), Expect = 5.9
Identities = 14/37 (37%), Positives = 20/37 (53%)
Frame = -3
Query: 356 FYTKASTRNMIKDIKIPVLFIQSDNGMVPAFSVPRNL 392
FY + D+K+PVLFI DN + A S +N+
Sbjct: 400 FYGTRIDSLVASDVKVPVLFILGDNDPICAVSEIKNI 290
>TC80489 similar to GP|9294341|dbj|BAB02238.1 contains similarity to unknown
protein~gb|AAF01552.1~gene_id:T6J22.18 {Arabidopsis
thaliana}, partial (68%)
Length = 1399
Score = 28.5 bits (62), Expect = 7.7
Identities = 13/27 (48%), Positives = 17/27 (62%)
Frame = +3
Query: 110 SISFKDNERLIKDEKHYGRIRVNKREK 136
SI FKD R+ D+K R V+KR+K
Sbjct: 1005 SIEFKDKARISTDQKEKSRSGVDKRDK 1085
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,463,142
Number of Sequences: 36976
Number of extensions: 249804
Number of successful extensions: 1416
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 1397
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1411
length of query: 602
length of database: 9,014,727
effective HSP length: 102
effective length of query: 500
effective length of database: 5,243,175
effective search space: 2621587500
effective search space used: 2621587500
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC124967.11


