

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124966.9 - phase: 0 /pseudo
(86 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
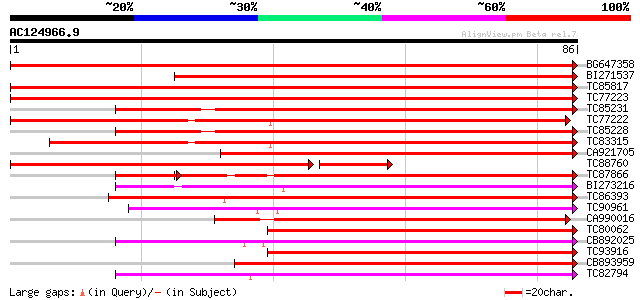
Score E
Sequences producing significant alignments: (bits) Value
BG647358 similar to GP|19911209|db glucosyltransferase-13 {Vigna... 159 1e-40
BI271537 similar to GP|19911209|db glucosyltransferase-13 {Vigna... 94 9e-21
TC85817 weakly similar to PIR|B85014|B85014 probable flavonol gl... 93 2e-20
TC77223 weakly similar to GP|19911209|dbj|BAB86931. glucosyltran... 88 5e-19
TC85231 similar to GP|13508844|emb|CAC35167. arbutin synthase {R... 86 2e-18
TC77222 weakly similar to GP|19911209|dbj|BAB86931. glucosyltran... 85 6e-18
TC85228 weakly similar to GP|19911209|dbj|BAB86931. glucosyltran... 84 1e-17
TC83315 weakly similar to GP|19911209|dbj|BAB86931. glucosyltran... 78 5e-16
CA921705 weakly similar to GP|13508844|em arbutin synthase {Rauv... 75 3e-15
TC88760 similar to GP|19911209|dbj|BAB86931. glucosyltransferase... 71 5e-15
TC87866 similar to GP|19911195|dbj|BAB86924. glucosyltransferase... 70 1e-14
BI273216 similar to GP|19911203|dbj glucosyltransferase-10 {Vign... 60 2e-10
TC86393 weakly similar to GP|10953887|gb|AAG25643.1 UDP-glucosyl... 59 3e-10
TC90961 similar to GP|20067056|gb|AAM09517.1 putative glucosyltr... 56 2e-09
CA990016 similar to GP|4140026|dbj flavonoid 3-O-galactosyl tran... 55 4e-09
TC80062 weakly similar to GP|4115534|dbj|BAA36410.1 UDP-glycose:... 55 5e-09
CB892025 similar to PIR|T06371|T06 probable UDP-glucuronosyltran... 54 8e-09
TC93916 weakly similar to GP|17065354|gb|AAL32831.1 putative pro... 54 8e-09
CB893959 weakly similar to GP|7385017|gb| UDP-glucose:salicylic ... 54 1e-08
TC82794 weakly similar to GP|3928543|dbj|BAA34687.1 UDP-glucose ... 54 1e-08
>BG647358 similar to GP|19911209|db glucosyltransferase-13 {Vigna angularis},
partial (33%)
Length = 738
Score = 159 bits (403), Expect = 1e-40
Identities = 79/86 (91%), Positives = 80/86 (92%)
Frame = +3
Query: 1 MEMGPIKAPTEEGSGNPSVYPVGPIIDTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGS 60
MEMGPI A TEEGSGNPSVYPVGPII TVT S DANGLECLSWLDKQQSCSVLYVSFGS
Sbjct: 210 MEMGPINALTEEGSGNPSVYPVGPIIQTVTGSVDDANGLECLSWLDKQQSCSVLYVSFGS 389
Query: 61 GGTLSHEQIVQLALGLELSNQKFLWV 86
GGTLSHEQIV+LALGLELSNQKFLWV
Sbjct: 390 GGTLSHEQIVELALGLELSNQKFLWV 467
>BI271537 similar to GP|19911209|db glucosyltransferase-13 {Vigna
angularis}, partial (34%)
Length = 652
Score = 94.0 bits (232), Expect = 9e-21
Identities = 46/61 (75%), Positives = 51/61 (83%)
Frame = +2
Query: 26 IDTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGSGGTLSHEQIVQLALGLELSNQKFLW 85
I T+ S +GLECL+WLDKQQ CSVLYVSFGSGGTLS EQIV+LALGLELSN+ FLW
Sbjct: 2 IPTIESSGDANHGLECLTWLDKQQPCSVLYVSFGSGGTLSQEQIVELALGLELSNKIFLW 181
Query: 86 V 86
V
Sbjct: 182V 184
>TC85817 weakly similar to PIR|B85014|B85014 probable flavonol
glucosyltransferase [imported] - Arabidopsis thaliana,
partial (83%)
Length = 1736
Score = 92.8 bits (229), Expect = 2e-20
Identities = 47/86 (54%), Positives = 57/86 (65%)
Frame = +3
Query: 1 MEMGPIKAPTEEGSGNPSVYPVGPIIDTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGS 60
+E GPIK +E G P YPVGP++ + E L WLD Q SVL+VSFGS
Sbjct: 789 LEPGPIKELLKEEPGKPKFYPVGPLVKREVEVGQIGPNSESLKWLDNQPHGSVLFVSFGS 968
Query: 61 GGTLSHEQIVQLALGLELSNQKFLWV 86
GGTLS +QIV+LALGLE+S Q+FLWV
Sbjct: 969 GGTLSSKQIVELALGLEMSEQRFLWV 1046
>TC77223 weakly similar to GP|19911209|dbj|BAB86931. glucosyltransferase-13
{Vigna angularis}, partial (50%)
Length = 1628
Score = 88.2 bits (217), Expect = 5e-19
Identities = 46/86 (53%), Positives = 55/86 (63%)
Frame = +3
Query: 1 MEMGPIKAPTEEGSGNPSVYPVGPIIDTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGS 60
+E IKA +G G +PVGPI T ++ + LECL WL Q SVLYVSFGS
Sbjct: 657 LESSAIKALELKGYGKIDFFPVGPITQTGLSNNDVGDELECLKWLKNQPQNSVLYVSFGS 836
Query: 61 GGTLSHEQIVQLALGLELSNQKFLWV 86
GGTLS QI +LA GLELS Q+F+WV
Sbjct: 837 GGTLSQTQINELAFGLELSGQRFIWV 914
>TC85231 similar to GP|13508844|emb|CAC35167. arbutin synthase {Rauvolfia
serpentina}, partial (34%)
Length = 1602
Score = 86.3 bits (212), Expect = 2e-18
Identities = 43/70 (61%), Positives = 52/70 (73%)
Frame = +3
Query: 17 PSVYPVGPIIDTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGSGGTLSHEQIVQLALGL 76
PSVYPVGP+I S+ +AN CL WL+ QQ SVL+VSFGSGGTLS +Q+ ++A GL
Sbjct: 744 PSVYPVGPMIRNE--SNNEANMSMCLRWLENQQPSSVLFVSFGSGGTLSQDQLNEIAFGL 917
Query: 77 ELSNQKFLWV 86
ELS KFLWV
Sbjct: 918 ELSGHKFLWV 947
>TC77222 weakly similar to GP|19911209|dbj|BAB86931. glucosyltransferase-13
{Vigna angularis}, partial (76%)
Length = 1643
Score = 84.7 bits (208), Expect = 6e-18
Identities = 45/87 (51%), Positives = 57/87 (64%), Gaps = 2/87 (2%)
Frame = +3
Query: 1 MEMGPIKAPTEEGSGNPSVYPVGPIIDTVTCSDRDANG--LECLSWLDKQQSCSVLYVSF 58
+E ++A ++G GN S +PVGPI + S+ D G ECL WL Q SVLYVSF
Sbjct: 666 LESSAVEALKQKGYGNISYFPVGPITQ-IGSSNNDVVGDEHECLKWLKNQPQNSVLYVSF 842
Query: 59 GSGGTLSHEQIVQLALGLELSNQKFLW 85
GSGGTLS QI ++A GLELS Q+F+W
Sbjct: 843 GSGGTLSQRQINEIAFGLELSGQRFIW 923
>TC85228 weakly similar to GP|19911209|dbj|BAB86931. glucosyltransferase-13
{Vigna angularis}, partial (31%)
Length = 3139
Score = 83.6 bits (205), Expect = 1e-17
Identities = 42/70 (60%), Positives = 51/70 (72%)
Frame = -1
Query: 17 PSVYPVGPIIDTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGSGGTLSHEQIVQLALGL 76
PSV+PVGPII S+ +AN CL WL+ Q SV++VSFGSGGTLS +Q+ +LA GL
Sbjct: 2092 PSVFPVGPIIRNE--SNNEANMSVCLRWLENQPPSSVIFVSFGSGGTLSQDQLNELAFGL 1919
Query: 77 ELSNQKFLWV 86
ELS KFLWV
Sbjct: 1918 ELSGHKFLWV 1889
>TC83315 weakly similar to GP|19911209|dbj|BAB86931. glucosyltransferase-13
{Vigna angularis}, partial (42%)
Length = 1048
Score = 78.2 bits (191), Expect = 5e-16
Identities = 44/82 (53%), Positives = 52/82 (62%), Gaps = 2/82 (2%)
Frame = +2
Query: 7 KAPTEEGSGNPSVYPVGPIIDTVTCSDRDANG--LECLSWLDKQQSCSVLYVSFGSGGTL 64
KA ++G G +PVG I + S+ D G ECL WL Q SVLYVSFGSGGTL
Sbjct: 638 KALEQKGYGKIGFFPVGTITQ-IGSSNNDVVGDEHECLKWLKNQPQNSVLYVSFGSGGTL 814
Query: 65 SHEQIVQLALGLELSNQKFLWV 86
S QI +LA GLELS Q+F+WV
Sbjct: 815 SQTQINELAFGLELSGQRFIWV 880
>CA921705 weakly similar to GP|13508844|em arbutin synthase {Rauvolfia
serpentina}, partial (34%)
Length = 787
Score = 75.5 bits (184), Expect = 3e-15
Identities = 35/54 (64%), Positives = 44/54 (80%)
Frame = -2
Query: 33 DRDANGLECLSWLDKQQSCSVLYVSFGSGGTLSHEQIVQLALGLELSNQKFLWV 86
D N +CLSWLD+Q+ SV++VSFGSGGT+S Q+ +LALGLELS+QKFLWV
Sbjct: 765 DNTQNESQCLSWLDEQKPNSVVFVSFGSGGTISQNQMNELALGLELSSQKFLWV 604
>TC88760 similar to GP|19911209|dbj|BAB86931. glucosyltransferase-13 {Vigna
angularis}, partial (34%)
Length = 862
Score = 70.9 bits (172), Expect(2) = 5e-15
Identities = 33/46 (71%), Positives = 35/46 (75%)
Frame = +3
Query: 1 MEMGPIKAPTEEGSGNPSVYPVGPIIDTVTCSDRDANGLECLSWLD 46
+EMGPI A EEGS NP VYPVGPII T T S DAN LECL+WLD
Sbjct: 690 LEMGPISAMKEEGSDNPPVYPVGPIIQTETSSGDDANALECLAWLD 827
Score = 24.3 bits (51), Expect(2) = 5e-15
Identities = 10/11 (90%), Positives = 10/11 (90%)
Frame = +2
Query: 48 QQSCSVLYVSF 58
QQ CSVLYVSF
Sbjct: 830 QQPCSVLYVSF 862
>TC87866 similar to GP|19911195|dbj|BAB86924. glucosyltransferase-6 {Vigna
angularis}, partial (12%)
Length = 960
Score = 70.5 bits (171), Expect(2) = 1e-14
Identities = 36/61 (59%), Positives = 44/61 (72%)
Frame = +3
Query: 26 IDTVTCSDRDANGLECLSWLDKQQSCSVLYVSFGSGGTLSHEQIVQLALGLELSNQKFLW 85
I+ C D + N + CL WL+ Q SVLYV FGSGGT+SHEQ+ +LA GLELS +KFLW
Sbjct: 759 INESNCED-NINSM-CLRWLENQPPNSVLYVCFGSGGTVSHEQLNELAFGLELSGKKFLW 932
Query: 86 V 86
V
Sbjct: 933 V 935
Score = 23.5 bits (49), Expect(2) = 1e-14
Identities = 9/10 (90%), Positives = 9/10 (90%)
Frame = +2
Query: 17 PSVYPVGPII 26
P VYPVGPII
Sbjct: 728 PCVYPVGPII 757
>BI273216 similar to GP|19911203|dbj glucosyltransferase-10 {Vigna
angularis}, partial (16%)
Length = 637
Score = 59.7 bits (143), Expect = 2e-10
Identities = 34/82 (41%), Positives = 45/82 (54%), Gaps = 12/82 (14%)
Frame = +3
Query: 17 PSVYPVGPIIDTVTCSDRDANGLE------------CLSWLDKQQSCSVLYVSFGSGGTL 64
P +Y +GP+ D + NG E CL WLD Q+ SVLYV+FGS +
Sbjct: 126 PKLYTLGPL-DLFLDKISENNGFESIQCNLWKEESECLKWLDSQEENSVLYVNFGSVIVM 302
Query: 65 SHEQIVQLALGLELSNQKFLWV 86
+ Q+V+LA GL S +KFLWV
Sbjct: 303 KYNQLVELAWGLANSKKKFLWV 368
>TC86393 weakly similar to GP|10953887|gb|AAG25643.1 UDP-glucosyltransferase
HRA25 {Phaseolus vulgaris}, partial (47%)
Length = 1332
Score = 58.9 bits (141), Expect = 3e-10
Identities = 31/72 (43%), Positives = 44/72 (61%), Gaps = 1/72 (1%)
Frame = +1
Query: 16 NPSVYPVGPIIDTVTC-SDRDANGLECLSWLDKQQSCSVLYVSFGSGGTLSHEQIVQLAL 74
+P P+GP+++ + S + L WLDKQ S SV+YVSFGS + Q +LAL
Sbjct: 670 SPKFLPIGPLMENDSNKSSFWQEDMTSLDWLDKQPSQSVVYVSFGSLAVMDQNQFNELAL 849
Query: 75 GLELSNQKFLWV 86
GL+L ++ FLWV
Sbjct: 850 GLDLLDKPFLWV 885
>TC90961 similar to GP|20067056|gb|AAM09517.1 putative glucosyltransferase
{Phaseolus lunatus}, partial (9%)
Length = 1130
Score = 56.2 bits (134), Expect = 2e-09
Identities = 30/78 (38%), Positives = 47/78 (59%), Gaps = 10/78 (12%)
Frame = +2
Query: 19 VYPVGPIIDTVTCSDRDA-------NGL---ECLSWLDKQQSCSVLYVSFGSGGTLSHEQ 68
V+ +GP++ + T ++ +G+ EC+ WLD + SVLY+SFGS T+S Q
Sbjct: 743 VWCIGPLLPSTTLKGSNSKYRAGKESGIALEECMEWLDLKDENSVLYISFGSQNTVSASQ 922
Query: 69 IVQLALGLELSNQKFLWV 86
++ LA GLE S + F+WV
Sbjct: 923 MMALAEGLEESEKLFIWV 976
>CA990016 similar to GP|4140026|dbj flavonoid 3-O-galactosyl transferase
{Vigna mungo}, partial (46%)
Length = 689
Score = 55.5 bits (132), Expect = 4e-09
Identities = 30/54 (55%), Positives = 35/54 (64%)
Frame = +2
Query: 32 SDRDANGLECLSWLDKQQSCSVLYVSFGSGGTLSHEQIVQLALGLELSNQKFLW 85
SDRD G CLSWLDKQ+S SV+YVSFG+ T ++V L LE S FLW
Sbjct: 314 SDRDETG--CLSWLDKQKSKSVIYVSFGTVATPPPNELVALTEALEESGFPFLW 469
>TC80062 weakly similar to GP|4115534|dbj|BAA36410.1 UDP-glycose:flavonoid
glycosyltransferase {Vigna mungo}, partial (42%)
Length = 1071
Score = 55.1 bits (131), Expect = 5e-09
Identities = 24/47 (51%), Positives = 33/47 (70%)
Frame = +2
Query: 40 ECLSWLDKQQSCSVLYVSFGSGGTLSHEQIVQLALGLELSNQKFLWV 86
ECLSWLD ++ SVLY+ FGS S +Q+ ++A G+E S KF+WV
Sbjct: 14 ECLSWLDSKEDNSVLYICFGSISHFSDKQLYEIASGIENSGHKFVWV 154
>CB892025 similar to PIR|T06371|T06 probable UDP-glucuronosyltransferase (EC
2.4.1.-) - garden pea, partial (32%)
Length = 708
Score = 54.3 bits (129), Expect = 8e-09
Identities = 29/80 (36%), Positives = 46/80 (57%), Gaps = 10/80 (12%)
Frame = +2
Query: 17 PSVYPVGPIIDTVTCSDR------DAN----GLECLSWLDKQQSCSVLYVSFGSGGTLSH 66
PS+Y +GP+ V S + D+N +CL WL+ ++ SV+YV+FGS +S
Sbjct: 389 PSLYTIGPLTSFVNQSPQNQFATLDSNLWKEDTKCLEWLESKEPASVVYVNFGSITIMSP 568
Query: 67 EQIVQLALGLELSNQKFLWV 86
E+ ++ A GL S + FLW+
Sbjct: 569 EKFLEFAWGLANSKKPFLWI 628
>TC93916 weakly similar to GP|17065354|gb|AAL32831.1 putative protein
{Arabidopsis thaliana}, partial (29%)
Length = 483
Score = 54.3 bits (129), Expect = 8e-09
Identities = 26/47 (55%), Positives = 34/47 (72%)
Frame = +1
Query: 40 ECLSWLDKQQSCSVLYVSFGSGGTLSHEQIVQLALGLELSNQKFLWV 86
E L+WL+ +Q+ SVLYVSFGS L H Q+V++A GLE S F+WV
Sbjct: 49 ELLNWLNSKQNESVLYVSFGSLTKLFHAQLVEIAHGLEKSGHNFIWV 189
>CB893959 weakly similar to GP|7385017|gb| UDP-glucose:salicylic acid
glucosyltransferase {Nicotiana tabacum}, partial (28%)
Length = 791
Score = 53.9 bits (128), Expect = 1e-08
Identities = 27/52 (51%), Positives = 34/52 (64%)
Frame = +3
Query: 35 DANGLECLSWLDKQQSCSVLYVSFGSGGTLSHEQIVQLALGLELSNQKFLWV 86
D N + WL+++ SV+YVSFGS LS EQI +LALGL S + FLWV
Sbjct: 360 DPNTESSIKWLNEKPKRSVVYVSFGSNARLSEEQIEELALGLNDSEKYFLWV 515
>TC82794 weakly similar to GP|3928543|dbj|BAA34687.1 UDP-glucose
glucosyltransferase {Arabidopsis thaliana}, partial
(59%)
Length = 1309
Score = 53.9 bits (128), Expect = 1e-08
Identities = 25/80 (31%), Positives = 46/80 (57%), Gaps = 10/80 (12%)
Frame = +1
Query: 17 PSVYPVGPIIDTVTCSDRD----------ANGLECLSWLDKQQSCSVLYVSFGSGGTLSH 66
PS+Y +GP+ + S ++ ++CL WL+ ++ SV+YV+FGS ++
Sbjct: 730 PSIYAIGPLTSFLNQSPQNNLASIGSNLWKEDMKCLEWLESKEQGSVVYVNFGSITVMTP 909
Query: 67 EQIVQLALGLELSNQKFLWV 86
+Q+++ A GL S + FLW+
Sbjct: 910 DQLLEFAWGLANSKKPFLWI 969
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.135 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,104,702
Number of Sequences: 36976
Number of extensions: 42815
Number of successful extensions: 219
Number of sequences better than 10.0: 107
Number of HSP's better than 10.0 without gapping: 213
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 214
length of query: 86
length of database: 9,014,727
effective HSP length: 62
effective length of query: 24
effective length of database: 6,722,215
effective search space: 161333160
effective search space used: 161333160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 51 (24.3 bits)
Medicago: description of AC124966.9


