

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124966.5 - phase: 0
(1309 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
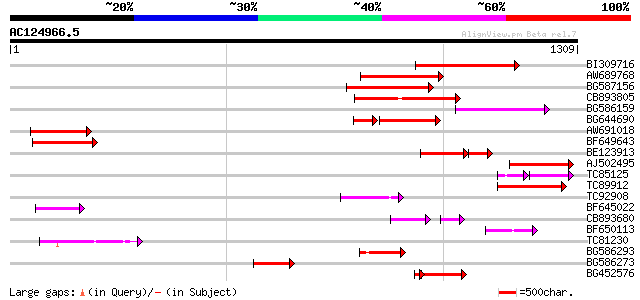
Score E
Sequences producing significant alignments: (bits) Value
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 261 1e-69
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 216 6e-56
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 209 7e-54
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 177 2e-44
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 169 8e-42
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 133 3e-41
AW691018 weakly similar to GP|2462935|em open reading frame 1 {B... 148 1e-35
BF649643 similar to PIR|H86486|H86 protein Ty1/copia-element pol... 143 5e-34
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 99 2e-30
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 129 7e-30
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 89 2e-29
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 117 2e-26
TC92908 similar to GP|10140704|gb|AAG13538.1 putative gag-pol po... 100 3e-21
BF645022 weakly similar to GP|8777581|dbj retroelement pol polyp... 93 6e-19
CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotia... 82 8e-19
BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vu... 91 3e-18
TC81230 91 4e-18
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 89 1e-17
BG586273 weakly similar to PIR|F86470|F8 probable retroelement p... 87 4e-17
BG452576 weakly similar to GP|12005223|gb|A reverse transcriptas... 64 2e-14
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 261 bits (668), Expect = 1e-69
Identities = 132/241 (54%), Positives = 171/241 (70%)
Frame = +2
Query: 936 VCKLNKSIYGLKQASRQWFSKFSTTLIQKGFRQSISDYSLFTYNCDQTTIFVLVYVDDII 995
VC+L KSIYGLKQASRQW+SK S +LI G+ QS SD+SLFT D + +LVYVDDI+
Sbjct: 20 VCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLLVYVDDIV 199
Query: 996 ITGNNENAISKIKKFLAQSFSIKDLGNLSYILGIEVSRSKKGIFLCQRKYTLDILSDSGM 1055
+ GN+ + I +K FL F IKDLG+L Y LG+EV+RSK+GI L QRKYTL++L DSG
Sbjct: 200 LAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEVARSKQGILLNQRKYTLELLEDSGN 379
Query: 1056 TGCRPSDFPMEQHLRLRPNDGTPLSDPTVYRRHVGRLLYLTVTRPDIQYAVNTLSQFMQS 1115
+ + P + L+L +D +D T YRR +G+L+YLT TRPDI +AV LSQF+
Sbjct: 380 LAVKSTLTPYDISLKLHNSDSPLYNDETQYRRLIGKLIYLTTTRPDISFAVQQLSQFVSK 559
Query: 1116 PYSSHFDAATQVLRYLKGSVGKGLFLSASSSINLVGYADSDWAGCPTTRRSTTGYFTMLG 1175
P H+ AA +VL+YLK + KGLF SA+S++ L +ADSDWA CPTTR+S TGY+ LG
Sbjct: 560 PQQVHYQAAIRVLQYLKTAPAKGLFYSATSNLKLSSFADSDWATCPTTRKSVTGYWVFLG 739
Query: 1176 S 1176
S
Sbjct: 740 S 742
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 216 bits (549), Expect = 6e-56
Identities = 103/191 (53%), Positives = 132/191 (68%)
Frame = +1
Query: 810 WRDAMAKEIQALESNNTWILCPLPEGKSAIGCKWIYKIKYHSDGSIDRYKARLVAKGYSQ 869
W AM E +AL N TW L PLP K AIGCKW+Y++K + DGS++++KARLVAKG+SQ
Sbjct: 97 WLQAMKTEYKALIDNKTWDLVPLPPHKKAIGCKWVYRVKENPDGSVNKFKARLVAKGFSQ 276
Query: 870 VQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAFLQGDLSEEVYMKLPPGFS 929
G DY +TF+PV K VT+RL+L+IA W + Q D+NNAFL G L EEVYM P GF
Sbjct: 277 TLGCDYTETFSPVIKPVTIRLILTIAITYKWEIQQIDINNAFLNGFLQEEVYMSQPQGFE 456
Query: 930 HKKKPCVCKLNKSIYGLKQASRQWFSKFSTTLIQKGFRQSISDYSLFTYNCDQTTIFVLV 989
K VCKLNKS+YGLKQA R W+ ++ IQ GF +S D SL YN + I++ +
Sbjct: 457 AANKSLVCKLNKSLYGLKQAPRAWYEXLTSAQIQFGFTKSRCDPSLLIYNQNGACIYLXI 636
Query: 990 YVDDIIITGNN 1000
YVDDI+ITG++
Sbjct: 637 YVDDILITGSS 669
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 209 bits (531), Expect = 7e-54
Identities = 101/200 (50%), Positives = 141/200 (70%), Gaps = 1/200 (0%)
Frame = -1
Query: 779 SNLSNSHRAFLINIIENKEPKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLPEGKSA 838
S +H AF++N+ EN P+SY +AM+ EW++++ E A+ N+TW LP+GK A
Sbjct: 615 SQYPEAHCAFMVNLNENHIPRSYEEAMEDKEWKESVGAEAGAMIKNDTWYESELPKGKKA 436
Query: 839 IGCKWIYKIKYHSDGSIDRYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIAAIK 898
+ +WI+ IKY +DGSI+R K RLVA+G++ G DY +TFAPVAKL T+R++LS+A
Sbjct: 435 VSSRWIFTIKYKADGSIERKKTRLVARGFTLTYGEDYIETFAPVAKLHTIRIVLSLAVNL 256
Query: 899 NWPLYQFDVNNAFLQGDLSEEVYMKLPPGFSH-KKKPCVCKLNKSIYGLKQASRQWFSKF 957
W L+Q DV NAFLQG+L +EVYM PPG H K+ V +L K+IYGLKQ+ R W++K
Sbjct: 255 GWGLWQMDVKNAFLQGELEDEVYMYPPPGLEHLVKRGNVLRLKKAIYGLKQSPRAWYNKL 76
Query: 958 STTLIQKGFRQSISDYSLFT 977
STTL +GFR+S D++LFT
Sbjct: 75 STTLNGRGFRKSELDHTLFT 16
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 177 bits (449), Expect = 2e-44
Identities = 92/253 (36%), Positives = 155/253 (60%), Gaps = 8/253 (3%)
Frame = +3
Query: 797 EPKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLPEGKSAIGCKWIYKIKYHSDGSID 856
+P ++ +A+KS +WR +M E++A E NNTW L L G IG KWI+K K + +G I+
Sbjct: 39 DPTTFEEAVKSEKWRASMNNEMEATERNNTWELTDLRSGAKTIGLKWIFKTKLNENGEIE 218
Query: 857 RYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAFL---- 912
+YKARLVAKGYSQ G+DY + FAPVA+ T+R+++++AA Q + +
Sbjct: 219 KYKARLVAKGYSQQYGVDYTEVFAPVARWDTIRMVIALAA-------QIKRDGVCIS*M* 377
Query: 913 QGDLSEEVYMKLPPGFSHK---KKPCVCKLNKSIYGLKQASRQWFSKFSTTLIQKGFRQS 969
+ E M+ +H+ ++ ++ +++YGLKQA R W+S+ ++GF +
Sbjct: 378 KAHSCMEN*MRKFLLINHRVM*RRVIS*RVKRALYGLKQAPRAWYSRIEAYFTKEGFEKC 557
Query: 970 ISDYSLFTYNCDQTTIFVL-VYVDDIIITGNNENAISKIKKFLAQSFSIKDLGNLSYILG 1028
+++LF + I ++ +YVDD+I GN+EN + KK + + F++ DLG + Y LG
Sbjct: 558 PYEHTLFVKLSEGGKILIISLYVDDLIFIGNDENMFEEFKKSMKKEFNMSDLGKMHYFLG 737
Query: 1029 IEVSRSKKGIFLC 1041
+EV++++KGI++C
Sbjct: 738 VEVTQNEKGIYIC 776
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 169 bits (427), Expect = 8e-42
Identities = 82/217 (37%), Positives = 129/217 (58%)
Frame = +1
Query: 1029 IEVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDFPMEQHLRLRPNDGTPLSDPTVYRRH 1088
+EV ++++GI++CQRKY D+L GM S P+ +L ++ D T Y++
Sbjct: 1 VEVIQNEEGIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIKDENGVKVDATKYKQI 180
Query: 1089 VGRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDAATQVLRYLKGSVGKGLFLSASSSIN 1148
VG L+YL TRPD+ Y ++ +S+FM P H A +VLRYL G++ G+ + S
Sbjct: 181 VGCLMYLAATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMYKRNGSEK 360
Query: 1149 LVGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEAEYRSLATLSSEL 1208
L Y DSD+AG R+ST+GY ML S +SW +KKQP ++ S+ +AE+ + A + +
Sbjct: 361 LEAYTDSDYAGDLDDRKSTSGYVFMLSSGAVSWSSKKQPVVTLSTTKAEFIAAAFCACQS 540
Query: 1209 QWLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPVFH 1245
W++ +L LG IT+YCD+ + I +++NPV H
Sbjct: 541 VWMRRVLEKLGYTQSGSITMYCDNNSTIKLSKNPVLH 651
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 133 bits (335), Expect(2) = 3e-41
Identities = 64/141 (45%), Positives = 96/141 (67%), Gaps = 1/141 (0%)
Frame = -2
Query: 855 IDRYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAFLQG 914
I R K++LV +GY+Q +GIDY + F+PVA++ +R+L++ AA + LYQ DV +AF+ G
Sbjct: 424 ITRNKSKLVVQGYNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKSAFING 245
Query: 915 DLSEEVYMKLPPGFSHKKKP-CVCKLNKSIYGLKQASRQWFSKFSTTLIQKGFRQSISDY 973
DL EEV++K PPGF + P V +LNK++YGLKQA R W+ + S L++ GF++ D
Sbjct: 244 DLKEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKRGKIDN 65
Query: 974 SLFTYNCDQTTIFVLVYVDDI 994
+LF + + + VYVDDI
Sbjct: 64 TLFLLKRE*ELLIIQVYVDDI 2
Score = 55.1 bits (131), Expect(2) = 3e-41
Identities = 20/56 (35%), Positives = 36/56 (63%)
Frame = -3
Query: 793 IENKEPKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLPEGKSAIGCKWIYKIK 848
I + EPK+ +A++ +W ++M +E+ E + W L P PEGK+ IG +W+++ K
Sbjct: 600 ISSIEPKNVKEALRDADWINSMQEELHQFERSKVWYLVPRPEGKTVIGTRWVFRNK 433
>AW691018 weakly similar to GP|2462935|em open reading frame 1 {Brassica
oleracea}, partial (12%)
Length = 639
Score = 148 bits (374), Expect = 1e-35
Identities = 67/141 (47%), Positives = 98/141 (68%), Gaps = 1/141 (0%)
Frame = +2
Query: 49 TTNPDPCVIHHSDNPSTVLVTPLLTGDNYGSWSRAVTMALRAKNKFGFVDGSLTIPKKKE 108
T+ PDP ++HHSD P LV L G NYG WSR++ ++L AKNK G +DG++ P +
Sbjct: 140 TSLPDPLLLHHSDTPGISLVNQPLNGGNYGEWSRSMLLSLSAKNKLGLIDGTVKAPSADD 319
Query: 109 DILT-WQRCNDLVASWILNSVSTEIRPSILYAETAEQIWSDLQDRFSQSNAPKIYQLKQS 167
L W+RCNDLV +WIL+S+ +I S+++++TA +WSDL DRFSQ + +IYQ++Q
Sbjct: 320 PKLPLWKRCNDLVLTWILHSIEPDIARSVIFSDTAAAVWSDLHDRFSQGDESRIYQIRQE 499
Query: 168 ISALKQEGMYVSLYFTQLKSL 188
IS +Q + +S Y+T+LKSL
Sbjct: 500 ISECRQGSLLISDYYTKLKSL 562
>BF649643 similar to PIR|H86486|H86 protein Ty1/copia-element polyprotein
[imported] - Arabidopsis thaliana, partial (2%)
Length = 608
Score = 143 bits (360), Expect = 5e-34
Identities = 64/152 (42%), Positives = 99/152 (65%), Gaps = 3/152 (1%)
Frame = +1
Query: 53 DPCVIHHSDNPSTVLVTPLLTGDNYGSWSRAVTMALRAKNKFGFVDGSLTIPKKKE---D 109
+P IH SD+P +LV L G NY SWSR++ AL AKNK GF++GS+ P + + +
Sbjct: 118 NPYYIHPSDHPGHLLVPTKLNGTNYSSWSRSMVHALTAKNKVGFINGSIKTPSEVDQPAE 297
Query: 110 ILTWQRCNDLVASWILNSVSTEIRPSILYAETAEQIWSDLQDRFSQSNAPKIYQLKQSIS 169
W RCN ++ SW+ +SV ++ +++A+TA Q+W D +D+FSQ N P IYQ+++S++
Sbjct: 298 YALWNRCNSMILSWLTHSVEPDLAKGVIHAKTACQVWEDFKDQFSQKNIPAIYQIQKSLA 477
Query: 170 ALKQEGMYVSLYFTQLKSLWDELNSIALVNPC 201
+L Q + S YFT++K LWDEL + + C
Sbjct: 478 SLSQGTVSASTYFTKIKGLWDELETYRTLPTC 573
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 99.4 bits (246), Expect(2) = 2e-30
Identities = 48/113 (42%), Positives = 75/113 (65%), Gaps = 1/113 (0%)
Frame = +1
Query: 948 QASRQWFSKFSTTLIQKGFRQSISDYSLFT-YNCDQTTIFVLVYVDDIIITGNNENAISK 1006
Q+ R WF +F+ + + G+ Q +D+++F ++ ++VYVDDI +TG++ I +
Sbjct: 1 QSPRDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILIVYVDDIFLTGDHGK*IKR 180
Query: 1007 IKKFLAQSFSIKDLGNLSYILGIEVSRSKKGIFLCQRKYTLDILSDSGMTGCR 1059
+K LA+ F IKDLGNL Y LG+EV+R KKG + QRKY LD+L ++ M GC+
Sbjct: 181 LKNLLAEEFEIKDLGNLKYFLGMEVARWKKGSSISQRKYVLDLLKETRMIGCK 339
Score = 52.8 bits (125), Expect(2) = 2e-30
Identities = 26/55 (47%), Positives = 37/55 (67%)
Frame = +2
Query: 1059 RPSDFPMEQHLRLRPNDGTPLSDPTVYRRHVGRLLYLTVTRPDIQYAVNTLSQFM 1113
+PS+ PM+ ++L D L D Y+R VG+L+YL+ TRPDI + V T+SQFM
Sbjct: 338 KPSETPMDATVKLGTLDNGTLVDKGRYQRLVGKLIYLSHTRPDISFVVCTMSQFM 502
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 129 bits (324), Expect = 7e-30
Identities = 61/148 (41%), Positives = 99/148 (66%)
Frame = +2
Query: 1155 SDWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEAEYRSLATLSSELQWLKYL 1214
SDWAG TR+ST+GY LG+ ISW +KKQP ++ S+AEAEY + + +++ WL+ +
Sbjct: 2 SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQPVVAFSTAEAEYIASTSCATQTVWLRRI 181
Query: 1215 LSDLGIDHPQPITIYCDSQAAIHIAENPVFHERTKHIEIDCHFVREKIKSGLIAPSYIRS 1274
L + + P IYCD+++AI +++NPVFH R+KHI+I H +RE I + Y +
Sbjct: 182 LEVMHHEQNTPTKIYCDNKSAIALSKNPVFHGRSKHIDIQFHKIRELIAEKEVVIEYCPT 361
Query: 1275 SDQLADIFTKPLGGDAYKRILGKLGVIE 1302
+++ADIFTKPL +++ ++ LG+++
Sbjct: 362 EEKIADIFTKPLKIESFYKLKKMLGMMK 445
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 89.4 bits (220), Expect(2) = 2e-29
Identities = 45/103 (43%), Positives = 61/103 (58%)
Frame = +3
Query: 1200 SLATLSSELQWLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPVFHERTKHIEIDCHFVR 1259
SL E W++ L+ +LG Q IT+YCDSQ+A+HIA NP FH RTKHI I HFVR
Sbjct: 228 SLPQACKEAIWMQRLMEELGHKQEQ-ITVYCDSQSALHIARNPAFHSRTKHIGIQYHFVR 404
Query: 1260 EKIKSGLIAPSYIRSSDQLADIFTKPLGGDAYKRILGKLGVIE 1302
E ++ G + I ++D LAD TK + D + G++E
Sbjct: 405 EVVEEGSVDMQKIHTNDNLADAMTKSINTDKFIWCRSSYGLLE 533
Score = 59.7 bits (143), Expect(2) = 2e-29
Identities = 29/73 (39%), Positives = 44/73 (59%)
Frame = +1
Query: 1126 QVLRYLKGSVGKGLFLSASSSINLVGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKTKK 1185
+++RY+KG+ G + S + + GY DSD+AG R+STTGY L +SW +K
Sbjct: 7 RIMRYIKGTSGVAVCFGGSE-LTVRGYVDSDFAGDHDKRKSTTGYVFTLAGGAVSWLSKL 183
Query: 1186 QPTISRSSAEAEY 1198
Q ++ S+ EAEY
Sbjct: 184 QTVVALSTTEAEY 222
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 117 bits (294), Expect = 2e-26
Identities = 64/160 (40%), Positives = 96/160 (60%), Gaps = 2/160 (1%)
Frame = +1
Query: 1127 VLRYLKGSVGKGLFLS--ASSSINLVGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKTK 1184
VL+YL S+ L + A L GY D+D+AG TR+S +G+ L ISWK
Sbjct: 16 VLKYLNESLKSSLKYTKAAQEEDALEGYVDADYAGNVDTRKSLSGFVFTLYGTTISWKAN 195
Query: 1185 KQPTISRSSAEAEYRSLATLSSELQWLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPVF 1244
+Q ++ S+ +AEY + + WLK ++ +LGI + + I+CDSQ+AIH+A + V+
Sbjct: 196 QQSVVTLSTTQAEYIAFVEGVKDAIWLKGMIGELGITQ-EYVKIHCDSQSAIHLANHQVY 372
Query: 1245 HERTKHIEIDCHFVREKIKSGLIAPSYIRSSDQLADIFTK 1284
HERTKHI+I HF+R+ I+S I + S + AD+FTK
Sbjct: 373 HERTKHIDIRLHFIRDMIESKEIVVEKMASEENPADVFTK 492
>TC92908 similar to GP|10140704|gb|AAG13538.1 putative gag-pol polyprotein
{Oryza sativa}, partial (1%)
Length = 638
Score = 100 bits (250), Expect = 3e-21
Identities = 63/144 (43%), Positives = 87/144 (59%)
Frame = +2
Query: 765 TSKINFPLENYLSLSNLSNSHRAFLINIIENKEPKSYSQAMKSVEWRDAMAKEIQALESN 824
+S ++ + +Y+ S S HRA L I N EPK+Y QA + EW AM +EIQ LE N
Sbjct: 185 SSSKSYHIFHYVGYS-FSAKHRASLAAITSNIEPKNYVQAAQ*QEWLAAMEQEIQVLEEN 361
Query: 825 NTWILCPLPEGKSAIGCKWIYKIKYHSDGSIDRYKARLVAKGYSQVQGIDYHDTFAPVAK 884
NT L PL EGK + C+ +YKI + ++G I++YKA+LVAK + QV+G D+ D K
Sbjct: 362 NTSTLEPLREGKKWVDCRPVYKIIHKANGEIEKYKAQLVAKDFVQVEGEDF*D-LCLSNK 538
Query: 885 LVTVRLLLSIAAIKNWPLYQFDVN 908
R LL+IAA K L+ DV+
Sbjct: 539 DDNCRCLLTIAAAKG*QLHLMDVS 610
>BF645022 weakly similar to GP|8777581|dbj retroelement pol polyprotein-like
{Arabidopsis thaliana}, partial (8%)
Length = 685
Score = 93.2 bits (230), Expect = 6e-19
Identities = 41/114 (35%), Positives = 66/114 (56%)
Frame = -2
Query: 60 SDNPSTVLVTPLLTGDNYGSWSRAVTMALRAKNKFGFVDGSLTIPKKKEDILTWQRCNDL 119
+DNP V+ L G NY WSRA+ + +AK K+GFV+G + P E + W+ +
Sbjct: 360 NDNPGNVITPIQLRGLNYDEWSRAIRTSFQAKRKYGFVEGKIPKPTTPEKLEDWKAVQSM 181
Query: 120 VASWILNSVSTEIRPSILYAETAEQIWSDLQDRFSQSNAPKIYQLKQSISALKQ 173
+ +W+LN++ +R ++ Y + AE +W+ L+ RF N +I QLK S+ KQ
Sbjct: 180 LIAWLLNTIEPSLRSTLSYYDDAESLWTHLKQRFCVVNGARICQLKASLGECKQ 19
>CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotiana tabacum},
partial (7%)
Length = 780
Score = 82.4 bits (202), Expect(2) = 8e-19
Identities = 46/93 (49%), Positives = 55/93 (58%)
Frame = -2
Query: 879 FAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAFLQGDLSEEVYMKLPPGFSHKKKPCVCK 938
F P+ KL T+ LLSI AI+N L DV AFL+GDL E++YM P GFS + V K
Sbjct: 554 FVPIVKLNTIMFLLSIVAIENLYLE*LDVKTAFLRGDLVEDIYMHQPEGFS*EVGKMVGK 375
Query: 939 LNKSIYGLKQASRQWFSKFSTTLIQKGFRQSIS 971
L KS+YGLKQ RQ +KGFR IS
Sbjct: 374 LKKSMYGLKQGPRQCI*SLKALCTRKGFRSEIS 276
Score = 30.8 bits (68), Expect(2) = 8e-19
Identities = 17/59 (28%), Positives = 33/59 (55%), Gaps = 2/59 (3%)
Frame = -3
Query: 994 IIITGNNENAISKIKKFLAQSFSIKDLGNLSYILGIEVSRSK-KGIF-LCQRKYTLDIL 1050
+++ G+N + I +K ++ +KDLG I+G+++ K KG+ L Q +Y +L
Sbjct: 271 LLVVGSNIDEIKNLKTRFSKEIDMKDLGPAKKIIGMQIMIDKQKGVL*LSQVEYITRVL 95
>BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vulgaris},
partial (13%)
Length = 494
Score = 90.9 bits (224), Expect = 3e-18
Identities = 50/123 (40%), Positives = 71/123 (57%), Gaps = 3/123 (2%)
Frame = +1
Query: 1099 RPDIQYAVNTLSQFMQSPYSSHFDAATQVLRYLKGSVGKGLFL---SASSSINLVGYADS 1155
RPDI Y+V+ +S+FM P H AA ++LRY++G++ GL + S L+ Y+DS
Sbjct: 121 RPDICYSVSVISKFMHDPRKPHLIAANRILRYVRGTMEYGLLFPYGAKSEVYELICYSDS 300
Query: 1156 DWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEAEYRSLATLSSELQWLKYLL 1215
DW G RRST+GY ISW TKKQP + SS EAEY + + + WL ++
Sbjct: 301 DWCG---DRRSTSGYVFKFNDAAISWCTKKQPITALSSYEAEYIAGTFATFQALWLDSVI 471
Query: 1216 SDL 1218
+L
Sbjct: 472 KEL 480
>TC81230
Length = 958
Score = 90.5 bits (223), Expect = 4e-18
Identities = 60/250 (24%), Positives = 116/250 (46%), Gaps = 12/250 (4%)
Frame = +1
Query: 68 VTPLLTGDNYGSWSRAVTMALRAKNKFGFVDGSLTIPKKKED---------ILTWQRCND 118
++ +L G NY W+ ++ L+ + + +V G P K +D + W N
Sbjct: 226 ISIILNGSNYNHWAESMCGFLKGRRLWRYVTGDKKCPTKGKDDTADAFADKLEEWDSKNH 405
Query: 119 LVASWILNSVSTEIRPSILYAETAEQIWSDLQDRFSQSNAPKIYQLKQSISALKQE-GMY 177
+ +W N+ I E A+++W L+ R++ S+ YQL + +S LKQ+ G
Sbjct: 406 QIITWFRNTSIPSIHMQFGRFENAKEVWDHLKQRYTISDLSHQYQLLKDLSNLKQQSGQP 585
Query: 178 VSLYFTQLKSLWDELNSIALVNPCI--CGNAKSILDQQNQDRAMEFLQGVHDRFSAVRSQ 235
V + Q++ +W++L S P + + K+ +N+ R ++FL + D + VR+
Sbjct: 586 VYEFLAQMEVIWNQLTS---CEPSLKDATDMKTYETHRNRVRLIQFLMALTDEYEPVRAS 756
Query: 236 ILLMDPFPSIQRIYNIVRQEEKQQEINFRPVPAVESAALQTSKAPYRTPGKRQRPFCEHC 295
L +P P+++ ++ EE + ++ P + A T+ A +P C HC
Sbjct: 757 SLHQNPLPTLENALPCLKSEETRLQL---VPPKADLAFAVTNNA--------TKP-CRHC 900
Query: 296 NRHGHTITTC 305
+ GH+ + C
Sbjct: 901 QKSGHSFSDC 930
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 88.6 bits (218), Expect = 1e-17
Identities = 48/106 (45%), Positives = 66/106 (61%)
Frame = +2
Query: 809 EWRDAMAKEIQALESNNTWILCPLPEGKSAIGCKWIYKIKYHSDGSIDRYKARLVAKGYS 868
E RD + + Q L+ L P G IG +WIYKIK + DG++ +YKARLVAKGY
Sbjct: 23 EGRDRIYYQKQTLK------LVKKPTGVKPIGLRWIYKIKRNEDGTLIKYKARLVAKGYV 184
Query: 869 QVQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAFLQG 914
+ QGID+ + FAPV ++ T+ LLL++AA ++ DV AFL G
Sbjct: 185 KQQGIDFDEVFAPVVRIETI*LLLALAATNGC*IHHIDVKIAFLNG 322
>BG586273 weakly similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 705
Score = 87.0 bits (214), Expect = 4e-17
Identities = 42/97 (43%), Positives = 65/97 (66%), Gaps = 1/97 (1%)
Frame = -2
Query: 562 AAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNMN-IQHKFDERSKPGIF 620
A YLIN++PT +LK ++P +VL PS + +RVFGCLC+ +++K + RS+ +F
Sbjct: 704 ACYLINRIPTRVLKDQAPFEVLNQRKPSLTYMRVFGCLCYVLVPGELRNKLEARSRKAMF 525
Query: 621 VGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQD 657
+GY QKGY+ YD + R++ VSRDV+F E Y++
Sbjct: 524 IGYSTTQKGYKCYDPEARRVLVSRDVKFIEERGYYEE 414
>BG452576 weakly similar to GP|12005223|gb|A reverse transcriptase-like protein
{Spiranthes hongkongensis}, partial (25%)
Length = 676
Score = 63.5 bits (153), Expect(2) = 2e-14
Identities = 38/104 (36%), Positives = 67/104 (63%)
Frame = +2
Query: 951 RQWFSKFSTTLIQKGFRQSISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENAISKIKKF 1010
R+ + K +TLI G++QS +D+S+F++ + TIF LVY+DDI+ ++ + +K
Sbjct: 236 RK*Y*KLLSTLISLGYKQSPNDHSIFSFG-RRFTIF-LVYLDDIVFARDDHSETQLVKSH 409
Query: 1011 LAQSFSIKDLGNLSYILGIEVSRSKKGIFLCQRKYTLDILSDSG 1054
L ++F I LG L Y+ G++++ S+ I Q KYT+++L +SG
Sbjct: 410 LDKNFIIIGLGTLHYV-GVKIA*SES*IIDDQCKYTIELLEESG 538
Score = 34.7 bits (78), Expect(2) = 2e-14
Identities = 17/27 (62%), Positives = 19/27 (69%), Gaps = 3/27 (11%)
Frame = +3
Query: 934 PC---VCKLNKSIYGLKQASRQWFSKF 957
PC VCK + SIYGLK A RQ +SKF
Sbjct: 150 PCLIAVCKFHNSIYGLKHAHRQ*YSKF 230
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.134 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 45,791,020
Number of Sequences: 36976
Number of extensions: 743835
Number of successful extensions: 5042
Number of sequences better than 10.0: 105
Number of HSP's better than 10.0 without gapping: 4771
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5006
length of query: 1309
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1201
effective length of database: 5,021,319
effective search space: 6030604119
effective search space used: 6030604119
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC124966.5


