

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124965.10 + phase: 0
(1569 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
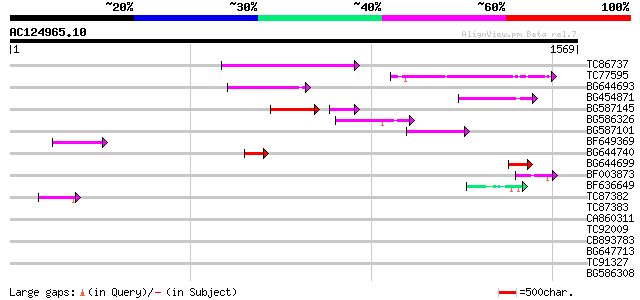
Score E
Sequences producing significant alignments: (bits) Value
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 251 2e-66
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 167 3e-41
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 162 1e-39
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 159 1e-38
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 106 1e-30
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 97 4e-20
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 75 2e-13
BF649369 73 1e-12
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 69 2e-11
BG644699 similar to PIR|T07863|T078 probable polyprotein - pinea... 64 6e-10
BF003873 similar to GP|14715222|em putative polyprotein {Cicer a... 62 2e-09
BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativ... 47 6e-05
TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulch... 44 6e-04
TC87383 similar to GP|19168656|emb|CAD26175. DNA-DIRECTED RNA PO... 41 0.004
CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product ... 40 0.005
TC92009 37 0.046
CB893783 weakly similar to GP|22830935|dbj hypothetical protein~... 37 0.046
BG647713 homologue to GP|15042313|gb| 232R {Chilo iridescent vir... 36 0.10
TC91327 similar to GP|22597168|gb|AAN03471.1 unknown protein {Gl... 36 0.10
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 36 0.10
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 251 bits (640), Expect = 2e-66
Identities = 148/396 (37%), Positives = 224/396 (56%), Gaps = 16/396 (4%)
Frame = +1
Query: 587 ATLLHTYREIFQIPKG--LPPNREL-SHEILL---KEGAQPVKVKPYRYPHSQKEQI--E 638
A +L + ++F K +P +R L H I L K+G P Y S++E + +
Sbjct: 337 AGVLEEFPDLFNPEKAYQVPASRGLLDHAIPLIPDKDGNDPPLPWGPLYGMSRQELLVLK 516
Query: 639 KMVQDMLEEGIIQPSLSPFSSPIILVKKKDGTWRCCTDYRALNAITIKDSFPMPTVDELL 698
K ++D+L++G I+ S S +P++ V+K G R C DYRALNAIT KD +P+P + E L
Sbjct: 517 KTLEDLLDKGFIKASGSAAGAPVLFVRKPGGGIRFCVDYRALNAITKKDRYPLPLISETL 696
Query: 699 DELHGAQYFTKLDLRSGYHQILLKPEDRQKTAFRTHQGHYEWLVMPFGLTSAPATFQRLM 758
+ GA++FTKLD+ + +H++ +K ED++KTAFRT G +EW+V PFGLT APATFQR +
Sbjct: 697 RRVAGARWFTKLDVVAAFHKMRIKDEDQEKTAFRTRYGLFEWIVCPFGLTGAPATFQRYI 876
Query: 759 NQLFQPLLRKCVLVFFDDILVYSP-SWHSHLKHVEIVFQILSQNVLYAKLSKCSFGITEV 817
N+ L V + DD+L+Y+ S H V V + L+ L KC F +T V
Sbjct: 877 NKTLHEFLDDFVTAYIDDVLIYTTGSKKDHEAQVRRVLRRLADAGLSLDPKKCEFSVTTV 1056
Query: 818 EYLGHVV-SGSGVAMDKIKVIAVLEWPTPTNLKQLRGFLGLTGYYRRFIRSYATIAGPLT 876
+Y+G ++ +G GV+ D +K+ A+ +W P ++K R FLG YY+ FI Y+ I PLT
Sbjct: 1057KYVGFILTAGKGVSCDPLKLAAIRDWLPPGSVKGARSFLGFCNYYKDFIPGYSEITEPLT 1236
Query: 877 NLLKKD-AFKWDESTAKAFDTLKQAITTAPVLVLPDFSQPFVLETDASGTGVGAVLSQ-- 933
L +KD F+W AF LK+ PVL + D +ETD SG +G VL+Q
Sbjct: 1237RLTRKDFPFRWGAEQEAAFTKLKRLFAEEPVLRMFDPEAVTTVETDCSGFALGGVLTQED 1416
Query: 934 ---GGHPIAYFSKKMASRMQLQSAYTREFYAITTAL 966
HP+A+ S++++ + +E A+ L
Sbjct: 1417GTGAAHPVAFHSQRLSPAEYNYPIHDKELLAVWACL 1524
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial (14%)
Length = 1708
Score = 167 bits (423), Expect = 3e-41
Identities = 137/484 (28%), Positives = 230/484 (47%), Gaps = 26/484 (5%)
Frame = +2
Query: 1054 DSHLQTIVQQCVNNTMDDINYMVKEGLLYWKHRLVIPMESN-------LIHQILKEYHDT 1106
D HL+ + +C +++ + + + R+ +P + L ++++E HD+
Sbjct: 50 DLHLKVQISECQLDSLKRLTF---------RGRIWVPGSDDEESPLNELRTKLVQESHDS 202
Query: 1107 PIGGHAGVTRTLARVTAQFYWPNMRQHIQKFIEACVTCQQAKSVNTTYAGLLQPLPIPEQ 1166
GH G TL V+ +F+WP Q +++F+ C C G L+PLP+P +
Sbjct: 203 TAAGHPGRNGTLEIVSRKFFWPGQSQTVRRFVRNCDVCGGIHIWRQAKRGFLKPLPVPNR 382
Query: 1167 VWDDVTMDFITGLPLSFG--FTVIMVVVDRLSKYAHFMPLKADYSNRTVAEAFVNYVVKL 1224
+ D++MDFIT LP + G + V+VDRLSK + A+ F++ +
Sbjct: 383 LHSDLSMDFITSLPPTRGRGSQYLWVIVDRLSKSVTLEEMDT-MEAEACAQRFLSCHYRF 559
Query: 1225 HGMPKSIISDRDKVFTSKFWQHLFQMQGTTLSMSTTYHPQTDGQSEAVNKCLEMYLRCFT 1284
HGMP+SI+SDR + +FW+ ++ G T +ST+YHPQTDG +E N+ ++ LR +
Sbjct: 560 HGMPQSIVSDRGSNWVGRFWREFCRLTGVTQLLSTSYHPQTDGGTERWNQEIQAVLRAYV 739
Query: 1285 FKNPKTWFKALTWVELWYNTFLHTSLGMSPFKALYG-REPPMLTRYSVNN--SDPTEVQQ 1341
+ W L V+L ++S+G +PF +G P+ T S+ Q
Sbjct: 740 CWSQDNWGDLLPTVQLALRNRHNSSIGATPFFVEHGYHVDPIPTVEDTGGVVSEGEAAAQ 919
Query: 1342 QLMDRDKLLVE-LKENLLRAQQVMKSNADKKRKDVS-FEVGDQVLVKLQPYRQSSVALRK 1399
L+ R K + ++ ++ AQQ +++A+K+R ++VGD+V + + Y+ R
Sbjct: 920 LLVKRMKDVTGFIQAEIVAAQQRSEASANKRRCPADRYQVGDKVWLNVSNYKSP----RP 1087
Query: 1400 NNKLGMRYFGPFTVIAKVGVVAYKLQLPETARIHPVFHVSQLK-----IFKGLTTEPYFP 1454
+ KL + + V V +L +P T ++P FHV L+ G P
Sbjct: 1088 SKKLDWLHH-KYEVTRFVTPHVVELNVPGT--VYPKFHVDLLRRAASDPLPGQEVVDPQP 1258
Query: 1455 LPLTTMEEGPVIQPEVILNVR--NIIRGDRKVEQLLVKWKDMQNSEATWE-----DKQEM 1507
P+ + + E IL R + RG R+ Q LVKWK +ATWE + E
Sbjct: 1259 PPIVDDDGEVEWEVEEILAARWHQVGRGRRR--QALVKWKGF--VDATWEAADAIRETEA 1426
Query: 1508 LDSY 1511
LD Y
Sbjct: 1427 LDRY 1438
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 162 bits (409), Expect = 1e-39
Identities = 89/235 (37%), Positives = 133/235 (55%), Gaps = 5/235 (2%)
Frame = +2
Query: 603 LPPNRELSHEILLKEGAQPVKVKPYRYPHSQKEQIEKMVQDMLEEGIIQPSLSPFSSPII 662
+PP ++ I L P+ + YR + + ++ ++D+LE+G IQPS+ P ++
Sbjct: 17 VPPEWKIDFGIDLLPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP*GVVVL 196
Query: 663 LVKKKDGTWRCCTDYRALNAITIKDSFPMPTVDELLDELHGAQYFTKLDLRSGYHQILLK 722
+KKKDG R DY LN + IK +P+P +DEL D L G+++F K+DLR G HQ +
Sbjct: 197 FLKKKDGFLRMSIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*HQHRVI 376
Query: 723 PEDRQKTAFRTHQGHYEWLVMPFGLTSAPATFQRLMNQLFQPLLRKCVLVFFDDILVYSP 782
ED KTAFR GHYE LVM FG T+ P F LMN++FQ L V+VF +DIL+YS
Sbjct: 377 GEDVPKTAFRIRYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDILIYSK 556
Query: 783 SWHSHLKHVEIVFQILSQNVLYAKLSKCSFG----ITEVEYLG-HVVSGSGVAMD 832
+ + H H+ + ++L + C + EV + HV+SG G+ +D
Sbjct: 557 NENEHENHLRLALKVLKD------IGLCQISYV*ILVEVGFFSLHVISGEGLKVD 703
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 159 bits (401), Expect = 1e-38
Identities = 92/224 (41%), Positives = 128/224 (57%), Gaps = 5/224 (2%)
Frame = +2
Query: 1241 SKFWQHLFQMQGTTLSMSTTYHPQTDGQSEAVNKCLEMYLRCFTFKNPKTWFKALTWVEL 1300
S FW+ LF++ GT L+MS+ YHP +DGQSEA+NK EMYLRC F +P W KA W E
Sbjct: 32 SNFWKQLFKLHGTILTMSSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFPWAEY 211
Query: 1301 WYNTFLHTSLGMSPFKALYGREPPMLTRYSVNNSDPTEVQQQLMDRDKLLVELKE----- 1355
WYNT + S M+PFKALYGR+ ML R ++ D ++Q QL R++LL +L+
Sbjct: 212 WYNTSYNISAAMTPFKALYGRDLSMLIRSKGSSKDTADLQSQLAQREELLSQLQSISTRL 391
Query: 1356 NLLRAQQVMKSNADKKRKDVSFEVGDQVLVKLQPYRQSSVALRKNNKLGMRYFGPFTVIA 1415
N L + +++++ A VS + + L+ ++ + T++
Sbjct: 392 NKL*SIKLIRNAAILSSSWVSTSL*NCNLINSLR*HCGNIKSSVHP----------TLVH 541
Query: 1416 KVGVVAYKLQLPETARIHPVFHVSQLKIFKGLTTEPYFPLPLTT 1459
AYKL LP TA++ P+FHVSQLK F G T PY LPLTT
Sbjct: 542 Y*QSAAYKLSLPSTAKVPPIFHVSQLKPFHGNIT*PYXRLPLTT 673
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 106 bits (264), Expect(2) = 1e-30
Identities = 50/136 (36%), Positives = 89/136 (64%)
Frame = +2
Query: 721 LKPEDRQKTAFRTHQGHYEWLVMPFGLTSAPATFQRLMNQLFQPLLRKCVLVFFDDILVY 780
+ P+D +KTAF T +G Y + VMPFGL +A +T+QRL+N++F L + V+ DD+LV
Sbjct: 2 MHPDDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVK 181
Query: 781 SPSWHSHLKHVEIVFQILSQNVLYAKLSKCSFGITEVEYLGHVVSGSGVAMDKIKVIAVL 840
S HL H++ F+ L + ++ +KC+FG+T E+LG++V+ G+ ++ ++ A+L
Sbjct: 182 SLRATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAIL 361
Query: 841 EWPTPTNLKQLRGFLG 856
+ P+P N ++++ G
Sbjct: 362 DLPSPKNSREVQRLTG 409
Score = 47.0 bits (110), Expect(2) = 1e-30
Identities = 30/89 (33%), Positives = 41/89 (45%), Gaps = 4/89 (4%)
Frame = +3
Query: 884 FKWDESTAKAFDTLKQAITTAPVLVLPDFSQPFVLETDASGTGVGAVLSQGGH----PIA 939
F WDE +AF+ LKQ +TT PVL P+ L S T V +VL + PI
Sbjct: 495 FVWDEKCEEAFEQLKQYLTTPPVLSKPEAGDTLSLYIAISSTAVSSVLIREDRGEQKPIF 674
Query: 940 YFSKKMASRMQLQSAYTREFYAITTALAK 968
Y SK+M + +A+ T+ K
Sbjct: 675 YTSKRMTDPETRYPTLEKMAFAVITSARK 761
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 97.4 bits (241), Expect = 4e-20
Identities = 75/248 (30%), Positives = 109/248 (43%), Gaps = 31/248 (12%)
Frame = +2
Query: 902 TTAPVLVLPDFSQPFVLETDASGTGVGAVLSQGGHPIAYFSKKMASRMQLQSAYTREFYA 961
T+AP+LVLP+ +V+ TDAS TG+G VL+Q IAY S+++ + E A
Sbjct: 8 TSAPILVLPELIT-YVVYTDASITGLGCVLTQHEKVIAYASRQLRKHEGNYPTHDLEMAA 184
Query: 962 ITTALAKFRHYLLGHKFILRTDQKSLKSLLDQSLQTPEQQAWLHKFIGFDFQIEYKPGKD 1021
+ AL +R YL G K + TD KSLK + Q Q+ W+ +D I Y PGK
Sbjct: 185 VVFALKIWRSYLYGAKVQIHTDHKSLKYIFTQPELNLRQRRWMEFVADYDLDITYYPGKA 364
Query: 1022 NQAADALSR-----------------------------VMSLSWSA-PEHDFLEQLKKEI 1051
N ADALSR SL A + D +++
Sbjct: 365 NLVADALSRRRVDVSAEREADDLDGMVRALRLNVLTKATESLGLEAVNQADLFTRIRLAQ 544
Query: 1052 GNDSHLQTIVQQCVNNTMDDINYM-VKEGLLYWKHRLVIPMESNLIHQILKEYHDTPIGG 1110
G D +LQ + Q D Y K+G + R+ +P + +L +I+ E H +
Sbjct: 545 GQDENLQKVAQN------DRTEYQTAKDGTILVNGRISVPNDRSLKEEIMSEAHKSRFSV 706
Query: 1111 HAGVTRTL 1118
H G R +
Sbjct: 707 HPGAPRCI 730
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 75.1 bits (183), Expect = 2e-13
Identities = 47/174 (27%), Positives = 88/174 (50%), Gaps = 1/174 (0%)
Frame = +2
Query: 1099 ILKEYHDTPIGGHAGVTRTLARVT-AQFYWPNMRQHIQKFIEACVTCQQAKSVNTTYAGL 1157
IL H + GH V++T++++ A F+WP M + FI C CQ+ +++ +
Sbjct: 104 ILFHCHGSNYAGHFAVSKTVSKIQQAGFWWPTMFKDAHSFISKCDPCQRQGNIS*RNE-M 280
Query: 1158 LQPLPIPEQVWDDVTMDFITGLPLSFGFTVIMVVVDRLSKYAHFMPLKADYSNRTVAEAF 1217
Q + +V+D +DF+ P S+ I+V VD +SK+ + + + V + F
Sbjct: 281 PQNFILEVEVFDVWGIDFMGPFPSSYNNKYILVAVDYVSKWVEAIASPTNDAT-VVVKMF 457
Query: 1218 VNYVVKLHGMPKSIISDRDKVFTSKFWQHLFQMQGTTLSMSTTYHPQTDGQSEA 1271
+ + G+P+ +ISD F +K ++ L + G ++T YHPQ +S++
Sbjct: 458 KSVIFPRFGVPRVVISDGGSHFINKVFEKLLKKNGVRHKVATAYHPQKAERSKS 619
>BF649369
Length = 631
Score = 72.8 bits (177), Expect = 1e-12
Identities = 45/152 (29%), Positives = 76/152 (49%)
Frame = +3
Query: 118 KLEFPRFNGKHVLDWIFKAEQYFGYYNTPDAERLIIASVHLEQEVVPWFQMVNRSRPFQS 177
K++ P F G + WI +AE YF NTPD R+ ++ + +E + WF ++ + S
Sbjct: 123 KVKLPLFEGDDPVAWITRAEIYFDVQNTPDDMRVKLSRLSMEGPTIHWFNLLMETEDDLS 302
Query: 178 WNDFTRALELDFGPSIYDCPRASLFKLQQNKSVNEYYLEFTALSNRVYGLSNDALIDCFV 237
+AL + + P L L+Q SV E+ F LS++V L + + F+
Sbjct: 303 REKLKKALIARYDGRRLENPFEELSTLRQIGSVEEFVEAFELLSSQVGRLPEEQYLGYFM 482
Query: 238 SGLKDELRRDVMLHTPISIVKAVSLAKLFEEK 269
SGLK +RR V P + ++ + +AK E++
Sbjct: 483 SGLKAHIRRRVRTLNPTTRMQMMRIAKDVEDE 578
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 68.6 bits (166), Expect = 2e-11
Identities = 29/65 (44%), Positives = 43/65 (65%)
Frame = -1
Query: 651 QPSLSPFSSPIILVKKKDGTWRCCTDYRALNAITIKDSFPMPTVDELLDELHGAQYFTKL 710
QPS+SP + ++ V+KKDG +R C DYR N +T K+ +P+P +D L D++ YF +
Sbjct: 262 QPSISP*GAALLFVRKKDGYFRMCIDYRQFNKVTTKNKYPLPRIDNLFDKIQEDCYF*NI 83
Query: 711 DLRSG 715
DLR G
Sbjct: 82 DLRLG 68
>BG644699 similar to PIR|T07863|T078 probable polyprotein - pineapple
retrotransposon dea1 (fragment), partial (5%)
Length = 231
Score = 63.5 bits (153), Expect = 6e-10
Identities = 31/67 (46%), Positives = 42/67 (62%), Gaps = 1/67 (1%)
Frame = +2
Query: 1381 DQVLVKLQPYRQSSVALRKNNKLGMRYFGPFTVIAKVGVVAYKLQLPE-TARIHPVFHVS 1439
+QVL+K+ P + K KL +RY GPF VI ++G VAY+L LP + +HPVFHVS
Sbjct: 2 EQVLLKVLPTERGDCRFGKRGKLSLRYIGPFEVIKRIGEVAYELALPPGLSGVHPVFHVS 181
Query: 1440 QLKIFKG 1446
K + G
Sbjct: 182 MFKRYHG 202
>BF003873 similar to GP|14715222|em putative polyprotein {Cicer arietinum},
partial (82%)
Length = 559
Score = 62.0 bits (149), Expect = 2e-09
Identities = 40/126 (31%), Positives = 62/126 (48%), Gaps = 10/126 (7%)
Frame = +2
Query: 1399 KNNKLGMRYFGPFTVIAKVGVVAYKLQL-PETARIHPVFHVSQLKIFKGLTTEPYFPLPL 1457
K+ KL +R+ GP+ + +VG VAY++ L P +H VFHVSQL+ Y P P
Sbjct: 20 KSKKLTVRFIGPYQISERVGTVAYRVGLPPHLLNLHDVFHVSQLR--------KYVPDPS 175
Query: 1458 TTMEEGPV-IQPEVILNVRNIIRGDRKVEQL--------LVKWKDMQNSEATWEDKQEML 1508
++ V ++ + + + DRKV+ L V W TWE + +M+
Sbjct: 176 HVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWDRANGESLTWELESKMV 355
Query: 1509 DSYPNL 1514
+SYP L
Sbjct: 356 ESYPEL 373
>BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativa}, partial
(4%)
Length = 653
Score = 47.0 bits (110), Expect = 6e-05
Identities = 49/186 (26%), Positives = 73/186 (38%), Gaps = 17/186 (9%)
Frame = +1
Query: 1265 TDGQSEAVNKCLEMYLRCFTFKNPKTWFKALTWVELWYNTFLHTSLGMSPFKALYGREPP 1324
+D Q+ +N LE +L FT + LTW E YNT H + G +PFK +Y
Sbjct: 7 SDEQAGLLNHTLETHLLYFTSEQQGV*NFFLTWAECLYNTNFHRTAGCTPFKVVY----- 171
Query: 1325 MLTRYSVNNSDPTEVQQQLMDRDKLLVELKENLLRAQQVMKSNADKKRKDVSFEVGDQVL 1384
+Q+ ++ RD L+ E L K SF G +
Sbjct: 172 ----------VVAHLQKFVVARD--LIYRNEGL-----------HKSST*TSFGRGTRAY 282
Query: 1385 VKL---------QPYRQSSVALRKNNKLGMRY--------FGPFTVIAKVGVVAYKLQLP 1427
L P R S+ ++ + +GP+ VI ++G VA+KL LP
Sbjct: 283 EALSRPAYETC*HPRRPLSIVYTRDRTYEWQVLPKYVA*CYGPYQVIKQIGSVAFKL*LP 462
Query: 1428 ETARIH 1433
E +IH
Sbjct: 463 EQHQIH 480
>TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulchellus},
partial (7%)
Length = 2304
Score = 43.5 bits (101), Expect = 6e-04
Identities = 27/124 (21%), Positives = 52/124 (41%), Gaps = 10/124 (8%)
Frame = +2
Query: 81 ATVVGSQSTFVGHTQSAVVGKQSTSSMSSQPFQVRHIKLEFPRFNGK----HVLDWIFKA 136
A + Q F G + +S + Q+ IK++ P F G LDW+
Sbjct: 506 ALLEAEQRRFEGDVSYSDSSSSRSSHSQRRQLQMNDIKVDIPDFEGNLQLDDFLDWLQTI 685
Query: 137 EQYFGYYNTPDAERLIIASVHLEQEVVPWFQMVNRSR------PFQSWNDFTRALELDFG 190
E+ F Y P+ +++ I + L++ + W++ + R R ++W+ + L +
Sbjct: 686 ERVFEYKEVPEEQKVKIVAAKLKKHALIWWENLKRRRKREGKSKIKTWDKMRQKLTRKYL 865
Query: 191 PSIY 194
P Y
Sbjct: 866 PPHY 877
>TC87383 similar to GP|19168656|emb|CAD26175. DNA-DIRECTED RNA POLYMERASE II
{Encephalitozoon cuniculi}, partial (0%)
Length = 1247
Score = 40.8 bits (94), Expect = 0.004
Identities = 34/154 (22%), Positives = 62/154 (40%), Gaps = 10/154 (6%)
Frame = -2
Query: 81 ATVVGSQSTFVGHTQSAVVGKQSTSSMSSQPFQVRHIKLEFPRFNGK----HVLDWIFKA 136
A + Q F H S+ +S + FQ+ IK + P F G +LDW+
Sbjct: 847 ALLEAQQKRFKDHVSSSDSLSSRSSRSQRREFQMNDIK*DIPDFEGNLQPDDLLDWLQIM 668
Query: 137 EQYFGYYNTPDAERLIIASVHLEQEVVPWFQMVNRSR------PFQSWNDFTRALELDFG 190
E+ F Y + +++ I + L++ W++ V R R ++W + L +
Sbjct: 667 ERLFKYKEVLEEQKVKIVAAKLKKLASIWWENVKRRRKREGKSKIKTWEKMRQKLTRKYL 488
Query: 191 PSIYDCPRASLFKLQQNKSVNEYYLEFTALSNRV 224
P Y + +L + S Y F+ N++
Sbjct: 487 PPHYYQDNYTQPQLSKKSS----YRHFSPTKNQI 398
>CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product {Drosophila
melanogaster}, partial (20%)
Length = 192
Score = 40.4 bits (93), Expect = 0.005
Identities = 25/60 (41%), Positives = 32/60 (52%), Gaps = 5/60 (8%)
Frame = +1
Query: 926 GVGAVLSQGG-----HPIAYFSKKMASRMQLQSAYTREFYAITTALAKFRHYLLGHKFIL 980
G+GA LSQ HPIAY S+ + + + + RE A A+ FRHYL G KF L
Sbjct: 13 GIGAGLSQKDEENHEHPIAYASRLLTAAERNYTVVERECLAAIWAIRNFRHYLHGPKFEL 192
>TC92009
Length = 974
Score = 37.4 bits (85), Expect = 0.046
Identities = 20/75 (26%), Positives = 34/75 (44%), Gaps = 1/75 (1%)
Frame = +2
Query: 117 IKLEFPRFNGKHVLDWIFKAEQYFGYYNTPDAERLIIASVHLEQE-VVPWFQMVNRSRPF 175
+K + +F G WI +AE +F ++L A + +E E + WF + P
Sbjct: 677 LKYKLSQFTGTDPAGWITRAEMFFADNEIHSCDKLQWAFMSMEDEKALLWFYSWCQENPD 856
Query: 176 QSWNDFTRALELDFG 190
W F+ A+ +FG
Sbjct: 857 ADWKSFSMAMIREFG 901
>CB893783 weakly similar to GP|22830935|dbj hypothetical protein~similar to
gag-pol polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 853
Score = 37.4 bits (85), Expect = 0.046
Identities = 49/205 (23%), Positives = 88/205 (42%), Gaps = 15/205 (7%)
Frame = +2
Query: 424 ISVSILLDGGSSESFIQPRIVHCLNLPIE---HTDKCNVLVGNGQHMKAEGVVRKLSLKV 480
I +++D GS E+ + +V L LP + H K L + ++ + S+
Sbjct: 254 IVCDVIIDSGSCENVVSNYMVEKLELPTKDHPHRYKLQWLKKGNEVRVSKCCLVSFSIGQ 433
Query: 481 QDIDITVPAYLLPVAGSDVILGAPWL----ASLGPHVADYSVSKLKFYMDGKFVTL---- 532
+ D V ++ + ++LG PW A H Y+ +K+ + K V L
Sbjct: 434 KYKD-NVWCDVISMDACHMLLGRPWQYDRHALYDGHANTYTF--VKYGVKIKLVPLPPNA 604
Query: 533 --QGESDNKPAVSQLNHFRRLQHMNAISELFTIQKIDPAVIEDNWEGMPVNIEPEMATLL 590
+G+ D KP VS ++ I ++ I +++ N E I+ E+ LL
Sbjct: 605 FDEGKKDFKPIVSLVSKEPFKVTTKDIQDMSLI-----LLVKSNEES---TIQKEVEHLL 760
Query: 591 HTYREIF--QIPKGLPPNRELSHEI 613
+ ++ +IP GLPP R++ H I
Sbjct: 761 VDFTDVVPSEIPSGLPPMRDIQHAI 835
>BG647713 homologue to GP|15042313|gb| 232R {Chilo iridescent virus}, partial
(1%)
Length = 726
Score = 36.2 bits (82), Expect = 0.10
Identities = 22/82 (26%), Positives = 41/82 (49%), Gaps = 8/82 (9%)
Frame = +2
Query: 100 GKQSTSSMSSQPFQ----VRHIKLEFPRFNGK----HVLDWIFKAEQYFGYYNTPDAERL 151
G S+SS SS+ + + IK++ P F GK +DW+ E+ F Y + +++
Sbjct: 269 GSDSSSSRSSRSHRRQTRMSKIKVDIPDF*GKLQPDEFVDWLQTIERVFKYKEVAEEQKV 448
Query: 152 IIASVHLEQEVVPWFQMVNRSR 173
I + L++ W++ + R R
Sbjct: 449 KIVAAKLKKHASIWWKNLKRKR 514
>TC91327 similar to GP|22597168|gb|AAN03471.1 unknown protein {Glycine max},
partial (70%)
Length = 738
Score = 36.2 bits (82), Expect = 0.10
Identities = 34/144 (23%), Positives = 59/144 (40%), Gaps = 5/144 (3%)
Frame = +1
Query: 208 KSVNEYYLEFTALSNRVYGLSNDALIDCFVSGLKDELRRDVMLHTPISIVKAVSLAKLFE 267
K V + +F + ++VY N+ ++ V E RD + +K++ + +
Sbjct: 112 KKVKKVLCKFPQIRDQVYDEKNN-IVTITVVCCSPEKIRDKICCKGCGAIKSIEIVEPPP 288
Query: 268 E-KIAANFNPKQPLKPYTPAQPHQHRAPFNPTRNDQNQTIEKAPNLPLLPTPPTRPMSHL 326
K PK+P+KP P +P + P NP + + + K P P P P P
Sbjct: 289 PPKPKEPEKPKEPVKPKEPEKPKEPEKPKNPEKPKEPEK-PKEPEKPKEPEKPKEPEKPK 465
Query: 327 QK----NPAIKRISPAEMQLRREK 346
+K P K P + + +EK
Sbjct: 466 EKPAPPPPEPKPEPPKQPEKPKEK 537
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 686
Score = 36.2 bits (82), Expect = 0.10
Identities = 16/52 (30%), Positives = 30/52 (56%)
Frame = -2
Query: 1225 HGMPKSIISDRDKVFTSKFWQHLFQMQGTTLSMSTTYHPQTDGQSEAVNKCL 1276
HG+P I++D F S ++ + L+ ++ +PQ++GQ+EA NK +
Sbjct: 685 HGLPYEIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKII 530
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 47,064,604
Number of Sequences: 36976
Number of extensions: 670901
Number of successful extensions: 3824
Number of sequences better than 10.0: 74
Number of HSP's better than 10.0 without gapping: 3707
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3802
length of query: 1569
length of database: 9,014,727
effective HSP length: 109
effective length of query: 1460
effective length of database: 4,984,343
effective search space: 7277140780
effective search space used: 7277140780
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Medicago: description of AC124965.10


