

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124963.22 + phase: 0 /pseudo
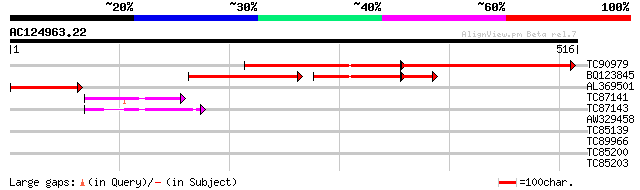
(516 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC90979 weakly similar to GP|9758056|dbj|BAB08519.1 folylpolyglu... 294 e-155
BQ123845 weakly similar to GP|17976703|em dihydrofolate syntheta... 199 e-101
AL369501 similar to GP|17976703|emb dihydrofolate synthetase /fo... 129 2e-30
TC87141 similar to GP|17976705|emb|CAC80839. dihydrofolate synth... 55 7e-08
TC87143 similar to GP|17976705|emb|CAC80839. dihydrofolate synth... 50 2e-06
AW329458 similar to GP|22136026|gb| unknown protein {Arabidopsis... 30 1.7
TC85139 homologue to GP|13540318|gb|AAK29410.1 S-adenosyl-L-meth... 30 2.9
TC89966 similar to PIR|B96768|B96768 protein enolase F2P9.10 [im... 29 3.8
TC85200 homologue to GP|10443981|gb|AAG17666.1 S-adenosylmethion... 29 5.0
TC85203 homologue to GP|14600068|gb|AAK71233.1 S-adenosylmethion... 28 6.5
>TC90979 weakly similar to GP|9758056|dbj|BAB08519.1 folylpolyglutamate
synthase-like protein {Arabidopsis thaliana}, partial
(20%)
Length = 1452
Score = 294 bits (752), Expect(2) = e-155
Identities = 154/159 (96%), Positives = 155/159 (96%)
Frame = +3
Query: 357 AHTKESAKALMNTIKMASPKARLAFVVAMASDKDHAGFAREILSDAYVKTVILTEAAIAG 416
AHTKESAKALMNTIKMASPKARLAFVVAMASDKDHAGFAREILS AYVKTVILTEAAIAG
Sbjct: 426 AHTKESAKALMNTIKMASPKARLAFVVAMASDKDHAGFAREILSXAYVKTVILTEAAIAG 605
Query: 417 AVTRTAPASLLRDSWIKASEELGTDICHDGMTEYRELFKEQPVSSESNLTDGKTILATES 476
AVTRT PASLLRDSWIKASE LG DICHDGMTEYRELFKEQPVSSESNLTDGKTILATES
Sbjct: 606 AVTRTTPASLLRDSWIKASEVLGIDICHDGMTEYRELFKEQPVSSESNLTDGKTILATES 785
Query: 477 SLKDCLRMANEILNRRRDEKGVIVITGSLHIVSSVLASL 515
SLKDCLR+ANEILNRRRDEKGVIVITGSLHIVSSVLASL
Sbjct: 786 SLKDCLRIANEILNRRRDEKGVIVITGSLHIVSSVLASL 902
Score = 272 bits (696), Expect(2) = e-155
Identities = 141/147 (95%), Positives = 142/147 (95%)
Frame = +2
Query: 214 YIIREKAVNMDSPVVSASDSGNNLAVKSFSILNGKPCQICDIEIQTVKDLKLAMDPDPRC 273
YIIREKAVNMDSPVVSASDSGNNLAVKSFSILNGKPCQICDIEIQTVKDLKLAMDPDPRC
Sbjct: 2 YIIREKAVNMDSPVVSASDSGNNLAVKSFSILNGKPCQICDIEIQTVKDLKLAMDPDPRC 181
Query: 274 ACGPVSCMI*SCKCLVLTNFKMQQLLLV*HSVFVI*VDGEFLMNL*GVG*SVHTSLEGVS 333
ACGPVSCMI*SCKCLVLTNFKMQQLLLV*HSVFVI* DGEFLMNL*GVG*SVHTSLEGVS
Sbjct: 182 ACGPVSCMI*SCKCLVLTNFKMQQLLLV*HSVFVI*-DGEFLMNL*GVG*SVHTSLEGVS 358
Query: 334 S*NLKKPRPWDFLEQQYCLI*SVAHTK 360
S*NLKKPRPWDFLEQQYCLI* H +
Sbjct: 359 S*NLKKPRPWDFLEQQYCLI**SPHQR 439
>BQ123845 weakly similar to GP|17976703|em dihydrofolate synthetase
/folylpolyglutamate synthetase {Arabidopsis thaliana},
partial (19%)
Length = 646
Score = 199 bits (507), Expect(3) = e-101
Identities = 101/104 (97%), Positives = 102/104 (97%)
Frame = +3
Query: 163 SGLAAAVITTVGEEHLAALGGSLESIAMAKAGIIKQGCPLVLGGPFLPHIEYIIREKAVN 222
SGLA VITTVGEEHLAALGGSLESIAMAKAGIIKQGCPLVLGGPFLPHIEYIIREKAVN
Sbjct: 3 SGLAVPVITTVGEEHLAALGGSLESIAMAKAGIIKQGCPLVLGGPFLPHIEYIIREKAVN 182
Query: 223 MDSPVVSASDSGNNLAVKSFSILNGKPCQICDIEIQTVKDLKLA 266
MDSPVVSASDSGNNLAVKSFSILNGKPCQICDIEIQTVKDLKL+
Sbjct: 183 MDSPVVSASDSGNNLAVKSFSILNGKPCQICDIEIQTVKDLKLS 314
Score = 145 bits (367), Expect(3) = e-101
Identities = 78/84 (92%), Positives = 79/84 (93%)
Frame = +1
Query: 277 PVSCMI*SCKCLVLTNFKMQQLLLV*HSVFVI*VDGEFLMNL*GVG*SVHTSLEGVSS*N 336
PVSCMI*SCKCLVLTNFKMQQLLLV*HSVFVI* DGEFLMNL*GVG*SVHTSLEGVSS*N
Sbjct: 313 PVSCMI*SCKCLVLTNFKMQQLLLV*HSVFVI*-DGEFLMNL*GVG*SVHTSLEGVSS*N 489
Query: 337 LKKPRPWDFLEQQYCLI*SVAHTK 360
LKKPRPWDFLEQQYCLI* H +
Sbjct: 490 LKKPRPWDFLEQQYCLI**SPHQR 561
Score = 62.4 bits (150), Expect(3) = e-101
Identities = 32/33 (96%), Positives = 32/33 (96%)
Frame = +2
Query: 357 AHTKESAKALMNTIKMASPKARLAFVVAMASDK 389
AHTKESAKALMNTIKMASPKARLAFV AMASDK
Sbjct: 548 AHTKESAKALMNTIKMASPKARLAFVGAMASDK 646
>AL369501 similar to GP|17976703|emb dihydrofolate synthetase
/folylpolyglutamate synthetase {Arabidopsis thaliana},
partial (10%)
Length = 470
Score = 129 bits (325), Expect = 2e-30
Identities = 63/66 (95%), Positives = 63/66 (95%)
Frame = +2
Query: 1 MRSVRTLCSHREDSEMKDLVDYIDSLKNYEKYGVPTGAGTDSNDGFDLGRMRRLLDRFGN 60
M SVRTLCS REDSEMKDLVDYIDSLKNYEK GVPTGAGTDSNDGFDLGRMRRLLDRFGN
Sbjct: 221 MCSVRTLCSLREDSEMKDLVDYIDSLKNYEKSGVPTGAGTDSNDGFDLGRMRRLLDRFGN 400
Query: 61 PHSKFK 66
PHSKFK
Sbjct: 401 PHSKFK 418
>TC87141 similar to GP|17976705|emb|CAC80839. dihydrofolate synthetase
/folylpolyglutamate synthetase {Arabidopsis thaliana},
partial (28%)
Length = 822
Score = 55.1 bits (131), Expect = 7e-08
Identities = 30/95 (31%), Positives = 50/95 (52%), Gaps = 3/95 (3%)
Frame = +3
Query: 69 GYSVGCYSSPHIQTIRERILGRSGDPVSAKLLNNL---FHRIKQDLDQAIKEENGCISHF 125
G+ G ++SPH+ IRER + K L + R+K+ D + + F
Sbjct: 528 GFRTGLFTSPHLIDIRERFRLDGVEICEEKFLAYFWWCYDRLKEKTDDKVPMP----TFF 695
Query: 126 EVFTAMAFILFADEKVDIAVIEAGLGGARDATNII 160
+AF +F++E+VD+ ++E GLGG DATN++
Sbjct: 696 HFLALLAFKIFSEEQVDVCIMEVGLGGKYDATNVV 800
>TC87143 similar to GP|17976705|emb|CAC80839. dihydrofolate synthetase
/folylpolyglutamate synthetase {Arabidopsis thaliana},
partial (41%)
Length = 920
Score = 50.4 bits (119), Expect = 2e-06
Identities = 31/110 (28%), Positives = 53/110 (48%)
Frame = +1
Query: 69 GYSVGCYSSPHIQTIRERILGRSGDPVSAKLLNNLFHRIKQDLDQAIKEENGCISHFEVF 128
G+ G ++SPH+ IRER F + D + + F
Sbjct: 286 GFRTGLFTSPHLIDIRER-----------------FRLDGKKTDDKVPMP----TFFHFL 402
Query: 129 TAMAFILFADEKVDIAVIEAGLGGARDATNIISSSGLAAAVITTVGEEHL 178
+AF +F++E+VD+ ++E GLGG DATN++ + IT++G +H+
Sbjct: 403 ALLAFKIFSEEQVDVCIMEVGLGGKYDATNVVHKPIVCG--ITSLGYDHM 546
>AW329458 similar to GP|22136026|gb| unknown protein {Arabidopsis thaliana},
partial (13%)
Length = 453
Score = 30.4 bits (67), Expect = 1.7
Identities = 17/53 (32%), Positives = 28/53 (52%), Gaps = 5/53 (9%)
Frame = +1
Query: 228 VSASDSGNNLAVKSFSILNGKPCQICDIEIQTVKDLKLAMDPD-----PRCAC 275
VS + +G+ K ++ +GK C+ D E+Q +K L +DP+ PR C
Sbjct: 109 VSEARTGSVTTYKRYADKDGKKCKRADGEVQRIKVSALKLDPEYWKNVPRRQC 267
>TC85139 homologue to GP|13540318|gb|AAK29410.1 S-adenosyl-L-methionine
synthetase {Elaeagnus umbellata}, complete
Length = 1542
Score = 29.6 bits (65), Expect = 2.9
Identities = 16/50 (32%), Positives = 21/50 (42%)
Frame = +2
Query: 226 PVVSASDSGNNLAVKSFSILNGKPCQICDIEIQTVKDLKLAMDPDPRCAC 275
P+ A S S+ G P ++CD V D L DPD + AC
Sbjct: 89 PLAPA*TMAETFLFTSESVNEGHPDKLCDQISDAVLDACLEQDPDSKVAC 238
>TC89966 similar to PIR|B96768|B96768 protein enolase F2P9.10 [imported] -
Arabidopsis thaliana, partial (46%)
Length = 680
Score = 29.3 bits (64), Expect = 3.8
Identities = 21/63 (33%), Positives = 32/63 (50%), Gaps = 4/63 (6%)
Frame = +3
Query: 187 SIAMAKAGIIKQGCPLVLGGPFLPHIEYIIREKAVNMDSPVVSA----SDSGNNLAVKSF 242
S+++ +AG +G PL HI+ I K + M P + S +GNNLA++ F
Sbjct: 306 SLSVCRAGAGAKGVPLY------KHIQEISGTKELVMPVPAFNVINGGSHAGNNLAMQEF 467
Query: 243 SIL 245
IL
Sbjct: 468 MIL 476
>TC85200 homologue to GP|10443981|gb|AAG17666.1 S-adenosylmethionine
synthetase {Brassica juncea}, partial (98%)
Length = 1689
Score = 28.9 bits (63), Expect = 5.0
Identities = 14/35 (40%), Positives = 18/35 (51%)
Frame = +1
Query: 241 SFSILNGKPCQICDIEIQTVKDLKLAMDPDPRCAC 275
S S+ G P ++CD V D L DPD + AC
Sbjct: 130 SESVNEGHPDKLCDQISDAVLDACLEQDPDSKVAC 234
>TC85203 homologue to GP|14600068|gb|AAK71233.1 S-adenosylmethionine
synthetase {Brassica juncea}, partial (27%)
Length = 635
Score = 28.5 bits (62), Expect = 6.5
Identities = 14/35 (40%), Positives = 18/35 (51%)
Frame = +3
Query: 241 SFSILNGKPCQICDIEIQTVKDLKLAMDPDPRCAC 275
S S+ G P ++CD V D L DPD + AC
Sbjct: 144 SESVNEGHPDKLCDQISDAVLDACLEQDPDNKVAC 248
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.327 0.140 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,627,438
Number of Sequences: 36976
Number of extensions: 168007
Number of successful extensions: 846
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 842
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 843
length of query: 516
length of database: 9,014,727
effective HSP length: 100
effective length of query: 416
effective length of database: 5,317,127
effective search space: 2211924832
effective search space used: 2211924832
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC124963.22


