

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124951.5 + phase: 0
(285 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
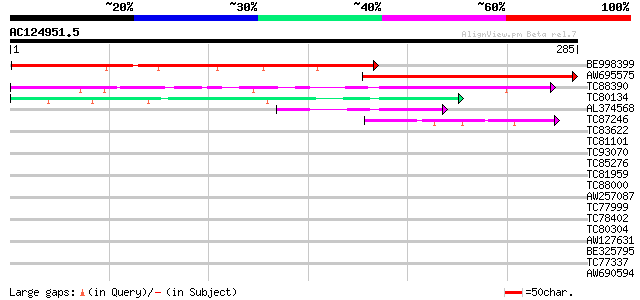
Score E
Sequences producing significant alignments: (bits) Value
BE998399 weakly similar to GP|18447465|gb| RE39957p {Drosophila ... 255 2e-68
AW695575 PIR|C85356|C85 glycine-rich protein [imported] - Arabid... 186 9e-48
TC88390 similar to GP|13272457|gb|AAK17167.1 unknown protein {Ar... 74 5e-14
TC80134 GP|7295572|gb|AAF50883.1| shakB gene product {Drosophila... 58 4e-09
AL374568 similar to GP|10177455|dbj gene_id:MQB2.9~pir||T01480~s... 55 4e-08
TC87246 similar to GP|15293109|gb|AAK93665.1 putative yeast pher... 41 5e-04
TC83622 similar to GP|21592533|gb|AAM64482.1 unknown {Arabidopsi... 34 0.056
TC81101 similar to GP|22136262|gb|AAM91209.1 unknown protein {Ar... 32 0.21
TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown prot... 32 0.28
TC85276 weakly similar to GP|22294973|dbj|BAC08801. ORF_ID:tlr12... 30 0.81
TC81959 similar to GP|6979444|gb|AAF34483.1| cytochrome oxidase ... 30 0.81
TC88000 weakly similar to GP|13486661|dbj|BAB39898. contains EST... 30 0.81
AW257087 homologue to GP|6683126|dbj KIAA0339 protein {Homo sapi... 30 1.1
TC77999 similar to SP|P29450|THIF_PEA Thioredoxin F-type chloro... 30 1.1
TC78402 weakly similar to PIR|B84853|B84853 hypothetical protein... 30 1.4
TC80304 30 1.4
AW127631 similar to PIR|T49904|T499 hypothetical protein T24H18.... 30 1.4
BE325795 similar to GP|22136848|gb| unknown protein {Arabidopsis... 30 1.4
TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Atel... 29 1.8
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 29 1.8
>BE998399 weakly similar to GP|18447465|gb| RE39957p {Drosophila
melanogaster}, partial (25%)
Length = 599
Score = 255 bits (651), Expect = 2e-68
Identities = 144/199 (72%), Positives = 157/199 (78%), Gaps = 15/199 (7%)
Frame = +3
Query: 2 QNQQQNEETSSTMDSFLCPSFSVYSSNNINDVAQQVTNENDNSHSQ--NDDFEFVAFRSH 59
QNQQQNEETSS+MDSFLCPSFS YSSNNINDV Q+VTNEND S++Q NDDFEFVAF +H
Sbjct: 3 QNQQQNEETSSSMDSFLCPSFSTYSSNNINDVVQKVTNENDTSNTQNDNDDFEFVAFHNH 182
Query: 60 RRNAADVSPIFNRD--ERRNSDAMDISISLKNLLIGDEKPKLNGGG--RRNSDAAEILYS 115
RRN DV PIFN D ERRNSDA +IS SLK LLIGDEK + + GG RRNSD AEI S
Sbjct: 183 RRN--DVFPIFNHDGNERRNSDAAEISNSLKKLLIGDEKQRNHDGGGRRRNSDVAEISNS 356
Query: 116 LKKLFIGNEKE---KWTSSEVEDDLDSIPAESYCFWTPKSS------SLIASPKTSPMNS 166
LKKLFIGNEKE + SSE+EDDLDSIPAE+YC WTP SS S I SP SPMNS
Sbjct: 357 LKKLFIGNEKEERNRVPSSELEDDLDSIPAETYCLWTPNSSPMNSPKSPITSPVASPMNS 536
Query: 167 TIKCKKSNSTGSSSNTSSS 185
KCKKSNSTGSSS++SSS
Sbjct: 537 LCKCKKSNSTGSSSSSSSS 593
>AW695575 PIR|C85356|C85 glycine-rich protein [imported] - Arabidopsis
thaliana, partial (9%)
Length = 555
Score = 186 bits (472), Expect = 9e-48
Identities = 92/108 (85%), Positives = 99/108 (91%)
Frame = -3
Query: 178 SSSNTSSSRWKFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGNGNVVGKKIPATEKKT 237
SSS++SSSRWKFLSLLRRSKSDGKESLN++TP KKENLK + G NV GKKIP TEKKT
Sbjct: 532 SSSSSSSSRWKFLSLLRRSKSDGKESLNLLTPVKKENLKKLNSGEKNVAGKKIPVTEKKT 353
Query: 238 PATVSAMEVFYGRKKETRVKSYLPYKKELIGFSVGFNANVGRGFPLHV 285
PATVSAMEVFY RKKE+RVKSYLPYKKELIGFSVGFNAN+GRGFPLHV
Sbjct: 352 PATVSAMEVFYRRKKESRVKSYLPYKKELIGFSVGFNANIGRGFPLHV 209
>TC88390 similar to GP|13272457|gb|AAK17167.1 unknown protein {Arabidopsis
thaliana}, partial (11%)
Length = 976
Score = 74.3 bits (181), Expect = 5e-14
Identities = 80/295 (27%), Positives = 121/295 (40%), Gaps = 21/295 (7%)
Frame = +2
Query: 1 MQNQQQNEETSSTMDSFLCPSFSVYSSNNINDVA-------QQVTNENDNSHS------Q 47
+Q +++ E S TM + S S ND +++ NEN++ H
Sbjct: 41 LQTKKKRETQSLTMQTVSTSPLSEVSRKFENDFFSKLKIEDEKIENENEDKHEVEEEDED 220
Query: 48 NDDFEFVAFRSHRRNAADVSPIFNRDERRNSDAMDISISLKNLLIGDEKPKLNGGGRRNS 107
+D+F FV F + + +F++ + R ++L D+ N G R+
Sbjct: 221 DDEFTFV-FADPKGSPISADDVFDKGQIRPI----FPFFGQDLHFTDDYD--NESGNRSP 379
Query: 108 DAAEILYSLKKLFI---GNEKEKWTSSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPM 164
++K F+ + K +S+ VE +YC W PK
Sbjct: 380 --------VRKFFVESPSSSKTATSSAAVEG--------TYCEWNPK------------- 472
Query: 165 NSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGNGN 224
K KSNSTGSS WK RS SDGK++ + P+K E ++G N
Sbjct: 473 ----KAVKSNSTGSSK-----LWKLRDPKLRSNSDGKDAFVFLNPSKVEKTSSSAGETKN 625
Query: 225 VVGKKIPATE-KKTPATVSAMEVFY----GRKKETRVKSYLPYKKELIGFSVGFN 274
V KK+ + KK SA E Y RK+ + KSYLPY++EL GF N
Sbjct: 626 DVVKKVKMVKGKKAAPAPSAHEKHYVMSKARKENDKRKSYLPYRQELFGFFASTN 790
>TC80134 GP|7295572|gb|AAF50883.1| shakB gene product {Drosophila
melanogaster}, partial (2%)
Length = 828
Score = 58.2 bits (139), Expect = 4e-09
Identities = 68/267 (25%), Positives = 102/267 (37%), Gaps = 39/267 (14%)
Frame = -3
Query: 1 MQNQQQNEETSSTMDSFL-------CPSFSVYSSNNINDVAQQVTNE------------- 40
MQ QQQ ++ ++ + PSFS YSS + ++A +V +E
Sbjct: 745 MQPQQQRDQGEASFSASSHNNHVSPSPSFSSYSSETLAEIAARVIDELRWDPHSDEDALY 566
Query: 41 ------------NDNSHSQNDDFEFVAFRSHRRNAADVSP--IFNRDERRNSDAMDISIS 86
NDN ++ +FEF N + VS IF + + + +
Sbjct: 565 QPWEDDNKLKTQNDNEKDEDSEFEFAVVSRDSTNFSVVSADDIFYNGQIK---PLYYPVF 395
Query: 87 LKNLLIGDEKPKLNGGGR-RNSDAAEILYSLKKLFIGNEKEKW----TSSEVEDDLDSIP 141
+NLL D+ ++ N L+ L E +S++ DL+ +
Sbjct: 394 DQNLLNDDDDGVVSSVSPVPNETTTRRRLPLRTLMFEESSEATASCSSSTDESIDLEGVA 215
Query: 142 AESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGK 201
SYC W P S I+ KKS+S GS SN RWK +LL RS SDGK
Sbjct: 214 EGSYCVWNPNSVG-------------IERKKSSSAGSGSN----RWKLRNLLLRSHSDGK 86
Query: 202 ESLNMVTPAKKENLKLNSGGNGNVVGK 228
+ ++ K S N GK
Sbjct: 85 DKQPVMFQIPKTTASKVSPAVENDCGK 5
>AL374568 similar to GP|10177455|dbj gene_id:MQB2.9~pir||T01480~similar to
unknown protein {Arabidopsis thaliana}, partial (8%)
Length = 520
Score = 54.7 bits (130), Expect = 4e-08
Identities = 33/87 (37%), Positives = 44/87 (49%), Gaps = 1/87 (1%)
Frame = +3
Query: 135 DDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLR 194
++ D + ++YC WTP CKKS+ST S+S RWKF LL
Sbjct: 141 EEFDDVAPDTYCVWTPP------------------CKKSSSTDSASK----RWKFRDLLL 254
Query: 195 RSKSDG-KESLNMVTPAKKENLKLNSG 220
RS SDG KESL ++P K+ +N G
Sbjct: 255 RSHSDGKKESLLFMSPGGKDRSAVNGG 335
>TC87246 similar to GP|15293109|gb|AAK93665.1 putative yeast pheromone
receptor protein AR781 {Arabidopsis thaliana}, partial
(26%)
Length = 1026
Score = 41.2 bits (95), Expect = 5e-04
Identities = 36/110 (32%), Positives = 56/110 (50%), Gaps = 12/110 (10%)
Frame = +1
Query: 179 SSNTSSSRWKFLS-LLRRSKSDGKESLNMVTPAKK----ENLKLNSGGNGNVV---GKKI 230
SS+ SS W+ LL RS S+G+ + P +K N KL+ G G+ +I
Sbjct: 640 SSSKSSRIWRLRDFLLFRSASEGRGTSK--DPFRKFPVSYNKKLSEEGKGSSSPFRSSEI 813
Query: 231 PATEKKTPATVSAMEVFYGRKK----ETRVKSYLPYKKELIGFSVGFNAN 276
P +K VSA E+ Y RKK + + +++LPYK+ ++G G +N
Sbjct: 814 PRPRRKDSG-VSAHELHYARKKAESEDLKKRTFLPYKQGILGRLAGLGSN 960
>TC83622 similar to GP|21592533|gb|AAM64482.1 unknown {Arabidopsis
thaliana}, partial (22%)
Length = 1066
Score = 34.3 bits (77), Expect = 0.056
Identities = 41/191 (21%), Positives = 80/191 (41%), Gaps = 38/191 (19%)
Frame = +3
Query: 123 NEKEKWTSSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNT 182
N + + E+ED++++ E+ + + + + T S+S SS+
Sbjct: 135 NNNKNNVACEIEDEMNNKNNENGNDCSEMENVKVDEMGLERIEITPLDSASSSRSSSAGR 314
Query: 183 SSSRWKFLSLLRRSKSDGKE-----SLNMVTPAK-KENLKLNSGGNGNVVGKKIPATEK- 235
SS RW FL RSKS+G+ S +P K K++ N + +K T++
Sbjct: 315 SSKRWVFLKDFLRSKSEGRSNNKFWSTISFSPTKDKKSSSNNQNLQAQISKEKTCETQRN 494
Query: 236 -KTPATVSAMEVFYGRK------------------------------KETRVKSYLPYKK 264
+ ++ S+ + +G+K +E R K++LPY++
Sbjct: 495 GSSSSSSSSSKGSWGKKMTGKPMNGVGKRRVPPSPHELHYKANRAQAEELRRKTFLPYRQ 674
Query: 265 ELIGFSVGFNA 275
L+G +GF++
Sbjct: 675 GLLG-CLGFSS 704
>TC81101 similar to GP|22136262|gb|AAM91209.1 unknown protein {Arabidopsis
thaliana}, partial (37%)
Length = 1188
Score = 32.3 bits (72), Expect = 0.21
Identities = 40/179 (22%), Positives = 69/179 (38%), Gaps = 8/179 (4%)
Frame = +3
Query: 25 YSSNNINDVAQQVTNENDNSHSQNDDFEFVAFRSHRRNAADVSPIFNRDERRNSDAMD-- 82
+S + + + N ++ + + DF+F + ++ +F+ ++
Sbjct: 363 FSQQGVIPIEKHPLRSNSSNMNSSIDFDFCVNETSELESSSADELFSHGIILPTELKKKK 542
Query: 83 ISISLKNLLIGDEKPK------LNGGGRRNSDAAEILYSLKKLFIGNEKEKWTSSEVEDD 136
I++ LK I P+ L G +NS KK+ + KE EV++
Sbjct: 543 INVPLKQPTIQPTPPQNFALPPLYANGSKNSS--------KKMITKDVKELNNIDEVDEK 698
Query: 137 LDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRR 195
S S FW K SS S + KSNSTGSS++ K S+L +
Sbjct: 699 HSS---NSKSFWGFKRSSSCGSGYGRSLCPLPLLSKSNSTGSSTSVKRMLSKEGSMLSK 866
>TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown protein
{Arabidopsis thaliana}, partial (19%)
Length = 679
Score = 32.0 bits (71), Expect = 0.28
Identities = 17/47 (36%), Positives = 30/47 (63%)
Frame = -3
Query: 157 ASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGKES 203
A+P +S +S+ S+S+ SSS++SSS K +S + S+S G+ +
Sbjct: 344 ANPSSSSSSSSSSSSSSSSSSSSSSSSSSSSKSISSISSSESYGERN 204
>TC85276 weakly similar to GP|22294973|dbj|BAC08801.
ORF_ID:tlr1249~hypothetical protein {Thermosynechococcus
elongatus BP-1}, partial (33%)
Length = 1315
Score = 30.4 bits (67), Expect = 0.81
Identities = 20/57 (35%), Positives = 28/57 (49%)
Frame = +2
Query: 159 PKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGKESLNMVTPAKKENL 215
P P S I SNS+ SSS++SSSR K + R K + + N K +N+
Sbjct: 929 PNQPPATSIIGSNGSNSSSSSSSSSSSRTKRMERRARQKMRSR-TTNEAQQQKDDNV 1096
>TC81959 similar to GP|6979444|gb|AAF34483.1| cytochrome oxidase subunit I
{Cordilura varipes}, partial (8%)
Length = 1487
Score = 30.4 bits (67), Expect = 0.81
Identities = 20/71 (28%), Positives = 34/71 (47%)
Frame = +3
Query: 120 FIGNEKEKWTSSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSS 179
+I + ++ SE E D+DS+P S T K S+ ++ S +N +S S S+
Sbjct: 306 YIAQNRSEFQESEFESDVDSVPLSS----TKKQKSIPSTSYISKLNHRPVSSRSLSQSST 473
Query: 180 SNTSSSRWKFL 190
+ S+ FL
Sbjct: 474 HFRNISQSDFL 506
>TC88000 weakly similar to GP|13486661|dbj|BAB39898. contains EST
AU081362(R10239)~unknown protein {Oryza sativa (japonica
cultivar-group)}, partial (20%)
Length = 1096
Score = 30.4 bits (67), Expect = 0.81
Identities = 16/40 (40%), Positives = 24/40 (60%)
Frame = +1
Query: 152 SSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLS 191
SSS ++ +SP S+ C S+S+ SSS + R +FLS
Sbjct: 454 SSSSPSNSPSSPYYSSASCSSSDSSSSSSKVVTRRRRFLS 573
Score = 27.7 bits (60), Expect = 5.3
Identities = 17/47 (36%), Positives = 29/47 (61%)
Frame = +1
Query: 154 SLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDG 200
++++S +SP NS S+++ SSS++SSS K ++ RR S G
Sbjct: 439 NIMSSSSSSPSNSPSSPYYSSASCSSSDSSSSSSKVVTRRRRFLSYG 579
>AW257087 homologue to GP|6683126|dbj KIAA0339 protein {Homo sapiens},
partial (2%)
Length = 466
Score = 30.0 bits (66), Expect = 1.1
Identities = 29/101 (28%), Positives = 40/101 (38%), Gaps = 12/101 (11%)
Frame = +1
Query: 125 KEKWTSSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKK------------ 172
K + EV+ D DS S T S S + P S +T K+
Sbjct: 130 KSASSKMEVDSDSDSDSDSSSSSSTSSSVSSVKKPAVSAAKATPAKKEESDSDSDSDSSS 309
Query: 173 SNSTGSSSNTSSSRWKFLSLLRRSKSDGKESLNMVTPAKKE 213
S+S+ SSS++SSS S S SD +E P K+
Sbjct: 310 SSSSSSSSSSSSSSSSSSSSSSSSNSDDEEEEEEAKPVVKK 432
>TC77999 similar to SP|P29450|THIF_PEA Thioredoxin F-type chloroplast
precursor (TRX-F). [Garden pea] {Pisum sativum},
complete
Length = 835
Score = 30.0 bits (66), Expect = 1.1
Identities = 25/61 (40%), Positives = 35/61 (56%), Gaps = 3/61 (4%)
Frame = +2
Query: 149 TPKSSSLIASPK---TSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGKESLN 205
TP + +L SPK T+ +ST K S GSSS+T+S F S + SKS G +SL+
Sbjct: 23 TPMALNLCTSPKWIGTTVFDSTSSSKLS--LGSSSSTAS----FSSSILNSKSVGLKSLS 184
Query: 206 M 206
+
Sbjct: 185 L 187
>TC78402 weakly similar to PIR|B84853|B84853 hypothetical protein At2g42370
[imported] - Arabidopsis thaliana, partial (14%)
Length = 1982
Score = 29.6 bits (65), Expect = 1.4
Identities = 24/58 (41%), Positives = 29/58 (49%), Gaps = 3/58 (5%)
Frame = -3
Query: 146 CF---WTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDG 200
CF W P S S S TS +S+ C S+S+ SSS T S FLSLL +G
Sbjct: 912 CFVEKWNPTSCS---SSSTSSSSSS-SCSCSSSSPSSSFTCSQSIPFLSLL*HGLKEG 751
>TC80304
Length = 787
Score = 29.6 bits (65), Expect = 1.4
Identities = 32/149 (21%), Positives = 63/149 (41%), Gaps = 7/149 (4%)
Frame = +2
Query: 101 GGGRRNS---DAAEILYSLKKLFIGNEKEKWTSSEVEDDLDSIPAESYCFWTPKSSSLIA 157
GGGR N + + L + ++ +G K+ S +ED L + + ++ A
Sbjct: 176 GGGRHNV*G*ETGQYLENQMQI-VGESVTKFYSEVMEDLLPPSSCATPVLPIGQYANAAA 352
Query: 158 SPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGKESLNMVTPAKKE---- 213
PK S S + K+N+ S+ ++S + + + N + K+
Sbjct: 353 IPKKSFQRSKKRMVKANTKRSTEDSS---------MNHDSGNSIKKSNFIWRPKQHVRSA 505
Query: 214 NLKLNSGGNGNVVGKKIPATEKKTPATVS 242
++KLN + N+ +KIPA + + +S
Sbjct: 506 DIKLNISDDENLKNRKIPAPKTASKVALS 592
>AW127631 similar to PIR|T49904|T499 hypothetical protein T24H18.70 -
Arabidopsis thaliana, partial (2%)
Length = 496
Score = 29.6 bits (65), Expect = 1.4
Identities = 30/116 (25%), Positives = 49/116 (41%), Gaps = 8/116 (6%)
Frame = +3
Query: 160 KTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGK--ESLNMVTPAKKENLKL 217
+TSP N + +CK+ NS S + SS +++ + +S N+V PA +
Sbjct: 129 QTSPTNESYRCKRRNSLEDSKSVSSCNKCNPAIITSELESARNSKSSNIVVPATDSHSSF 308
Query: 218 NSGGNG-NVVGKKIPATEKK-----TPATVSAMEVFYGRKKETRVKSYLPYKKELI 267
S V+ P +KK +P + +V K+ + S KKELI
Sbjct: 309 QSETKSKGVISWLFPRLKKKHKNENSPNRAESEDVSQ-VLKDIGIMSIEALKKELI 473
>BE325795 similar to GP|22136848|gb| unknown protein {Arabidopsis thaliana},
partial (6%)
Length = 676
Score = 29.6 bits (65), Expect = 1.4
Identities = 21/62 (33%), Positives = 29/62 (45%)
Frame = +1
Query: 124 EKEKWTSSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTS 183
+K T + L + P Y F +S+ KT +NST K +NST SS+N S
Sbjct: 493 QKPNKTIKRFKTHL*NHPHRHYYFAQKLNSTTF---KTKKLNSTSKASTTNSTKSSTNKS 663
Query: 184 SS 185
S
Sbjct: 664 KS 669
>TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Ateline
herpesvirus 3}, partial (8%)
Length = 986
Score = 29.3 bits (64), Expect = 1.8
Identities = 15/36 (41%), Positives = 23/36 (63%)
Frame = -2
Query: 152 SSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRW 187
SSS S ++P +S+ S+S+ SSS++SSS W
Sbjct: 730 SSSSSLSSSSTPSSSSSSPPSSSSSSSSSSSSSSSW 623
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 29.3 bits (64), Expect = 1.8
Identities = 15/34 (44%), Positives = 23/34 (67%)
Frame = -3
Query: 152 SSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSS 185
SSS +S +S +S+ C S+S+ SSS++SSS
Sbjct: 226 SSSSSSSSSSSSSSSSSSCSSSSSSSSSSSSSSS 125
Score = 28.9 bits (63), Expect = 2.4
Identities = 21/67 (31%), Positives = 34/67 (50%)
Frame = -3
Query: 149 TPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGKESLNMVT 208
+P SSS +S +S +S+ S+S+ SSS++SSS S S S S + +
Sbjct: 241 SPSSSSSSSSSSSSSSSSSSSSSCSSSSSSSSSSSSSS----SCSSSSSSSCSSSSSSTS 74
Query: 209 PAKKENL 215
P+ +L
Sbjct: 73 PSSSSSL 53
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.309 0.126 0.351
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,267,874
Number of Sequences: 36976
Number of extensions: 114247
Number of successful extensions: 979
Number of sequences better than 10.0: 65
Number of HSP's better than 10.0 without gapping: 827
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 893
length of query: 285
length of database: 9,014,727
effective HSP length: 95
effective length of query: 190
effective length of database: 5,502,007
effective search space: 1045381330
effective search space used: 1045381330
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC124951.5


