

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124217.5 + phase: 0 /pseudo
(1307 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
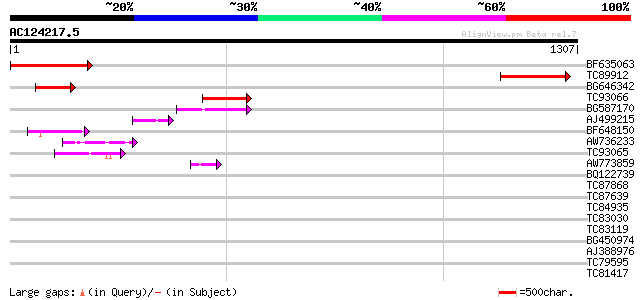
Score E
Sequences producing significant alignments: (bits) Value
BF635063 weakly similar to PIR|F84486|F84 probable retroelement ... 293 4e-79
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 259 6e-69
BG646342 weakly similar to PIR|F84486|F84 probable retroelement ... 125 8e-29
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 115 8e-26
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 70 4e-12
AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {V... 56 8e-08
BF648150 similar to GP|14586969|gb| pol polyprotein {Citrus x pa... 49 1e-05
AW736233 GP|21105451|gb small nuclear ribonucleoprotein D1 {Dani... 47 4e-05
TC93065 44 3e-04
AW773859 44 4e-04
BQ122739 38 0.022
TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 37 0.050
TC87639 GP|9663153|emb|CAC01132.1 transport-secretion protein 2.... 37 0.050
TC84935 similar to PIR|G96631|G96631 probable RNA-binding protei... 35 0.25
TC83030 weakly similar to GP|18855061|gb|AAL79753.1 putative RNA... 34 0.42
TC83119 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 34 0.42
BG450974 similar to PIR|T05112|T05 splicing factor 9G8-like SR p... 33 0.55
AJ388976 similar to PIR|E84638|E84 probable RSZp22 splicing fact... 33 0.94
TC79595 similar to GP|3850823|emb|CAA77136.1 U2 snRNP auxiliary ... 32 2.1
TC81417 weakly similar to GP|10998142|dbj|BAB03113. gene_id:MEC1... 31 2.7
>BF635063 weakly similar to PIR|F84486|F84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(4%)
Length = 677
Score = 293 bits (749), Expect = 4e-79
Identities = 150/189 (79%), Positives = 167/189 (87%)
Frame = -2
Query: 2 MGSKWDIEKFTGSNDFGLWKVKMQAVLTQQKCVEALKGEAAMPATLTQEEKREMIDKAKS 61
MGSK DIEKFTG NDFGLWKVKM+AVL QQKC +ALKGE ++P T+++ EK EM+DKA+S
Sbjct: 568 MGSKRDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEVSLPVTMSRAEKTEMVDKARS 389
Query: 62 AIVLCLGDKVLRDVAREATAASM*AKLESLYMTKSLAHRQLLKQQLYSFKMVESISISEQ 121
AIVLCLGDKVLR+VA+E TAASM AKL SLYMTKSLAHRQ LKQQLYSF+MVES +I EQ
Sbjct: 388 AIVLCLGDKVLREVAKERTAASMWAKL*SLYMTKSLAHRQFLKQQLYSFRMVESKAIMEQ 209
Query: 122 LTEFNKILVDLANIEVNTEDEDKALLLLCSLPKSFEHFKDTILYGKEGTTTLEEVQAALR 181
LTEFNKIL DL NIEV EDE+KA+LLLC+LPKSFE FKDT+LYGKEGT TLEEVQAALR
Sbjct: 208 LTEFNKILDDLENIEVQLEDEEKAILLLCALPKSFESFKDTMLYGKEGTVTLEEVQAALR 29
Query: 182 TKELTKFKD 190
TKELTK D
Sbjct: 28 TKELTKSND 2
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 259 bits (661), Expect = 6e-69
Identities = 137/164 (83%), Positives = 143/164 (86%), Gaps = 3/164 (1%)
Frame = +3
Query: 1131 SFEVGVEVFEWVFEGRFEVHKSSSRERRFGGVC*C---GQCGH*KILIGFCVYSLWHGY* 1187
SFEVGVEVFE VFE +FEVHKSSSR RRFGGVC*C GQCGH KI IGFCVYSLWH Y*
Sbjct: 3 SFEVGVEVFE*VFEEQFEVHKSSSRGRRFGGVC*CRLCGQCGHKKISIGFCVYSLWHDY* 182
Query: 1188 LEGKSTIRGCIIYHSSGVHCPC*RGERGHMVERDDW*VRNYSRMCEDTL**SKCNSLGKS 1247
LEGKSTIRG II +SSGVHC C RGER HMVER DW*VRNYSR+CEDTL**SKC+SLG+S
Sbjct: 183 LEGKSTIRGDIINNSSGVHCLCRRGERCHMVERYDW*VRNYSRICEDTL**SKCHSLGES 362
Query: 1248 SSVS*EDKAH*HSLALCQRHD*IKRDCG*KGGIGRESGECVHQV 1291
SSVS*ED AH*HSLAL RHD*IKRDCG K GIGRESG CV+QV
Sbjct: 363 SSVS*ED*AH*HSLALY*RHD*IKRDCGGKNGIGRESGGCVYQV 494
>BG646342 weakly similar to PIR|F84486|F84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(4%)
Length = 599
Score = 125 bits (315), Expect = 8e-29
Identities = 69/94 (73%), Positives = 78/94 (82%)
Frame = +2
Query: 59 AKSAIVLCLGDKVLRDVAREATAASM*AKLESLYMTKSLAHRQLLKQQLYSFKMVESISI 118
A+SAIVLCLGDKVLR+VA+E TA SM AKLE LYMTKSLAHRQ LKQQLYSFKMVES +I
Sbjct: 2 ARSAIVLCLGDKVLREVAKEPTATSMCAKLEYLYMTKSLAHRQFLKQQLYSFKMVESKAI 181
Query: 119 SEQLTEFNKILVDLANIEVNTEDEDKALLLLCSL 152
+E L EFNKI+ DL NIEV+ ED AL++ C L
Sbjct: 182 TELLVEFNKIIGDLENIEVHLEDAG-ALMVWCCL 280
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 115 bits (289), Expect = 8e-26
Identities = 53/114 (46%), Positives = 77/114 (67%), Gaps = 1/114 (0%)
Frame = +1
Query: 444 LDKLEFCEHCIL-GKQHRVKFGSGMHHSSRLFEYVHSDLLGPSKTPTHGGGSYFLSIIDD 502
+DKLEFC+H + G + +V F + H + + +Y+HSDL GPSK ++GG Y ++IIDD
Sbjct: 4 IDKLEFCKHLLFFGNRKKVSFSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDD 183
Query: 503 YSRRVWVFVLKKKSDTF*KFKE*HTLIENQMGTKLKGLRTDNGLEFVSEQFNDF 556
+ R+VWV+ L+ K++TF FK+ L+E Q G +K L TDN LEF S FN+F
Sbjct: 184 FPRKVWVYFLRYKNETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEF 345
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 70.5 bits (171), Expect = 4e-12
Identities = 52/175 (29%), Positives = 79/175 (44%), Gaps = 2/175 (1%)
Frame = -3
Query: 385 KGSKMNGLYILDGSIVIGN--ASVASVVPHNNSELWHLRLGHVSERGLVELAKQGLLGKD 442
+G LY+L+ + N S S N LWH RLGH R L L G++ ++
Sbjct: 686 EGVTKGDLYMLEKLDPVSNYKCSFTSSSSLNKDALWHARLGHPHGRAL-NLMLPGVVFEN 510
Query: 443 KLDKLEFCEHCILGKQHRVKFGSGMHHSSRLFEYVHSDLLGPSKTPTHGGGSYFLSIIDD 502
K CE CILGK + F F+ +++D L + + + YF++ ID+
Sbjct: 509 K-----NCEACILGKHCKNVFPRTSTVYENCFDLIYTD-LWTAPSLSRDNHKYFVTFIDE 348
Query: 503 YSRRVWVFVLKKKSDTF*KFKE*HTLIENQMGTKLKGLRTDNGLEFVSEQFNDFL 557
S+ W+ ++ K FK + N K+K LR+DNG E+ S F L
Sbjct: 347 KSKYTWLTLIPSKDRVIDAFKNFQAYVTNHYHAKIKILRSDNGGEYTSYAFKSHL 183
>AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {Vitis
vinifera}, partial (18%)
Length = 567
Score = 56.2 bits (134), Expect = 8e-08
Identities = 30/94 (31%), Positives = 51/94 (53%)
Frame = +3
Query: 283 KSWVLDSGCSYHMCPRKEYFETLTLKEGGVVRLGNNKACKVQGMGNVRLKMFDGREFLLR 342
K W++DSGC++HM ++ F+ L VR+ N +V+G+G V +K G + +
Sbjct: 102 KYWLIDSGCTHHMTHDRDLFKELNKSTISKVRMLNGAHIEVEGIGTVLVKSHSGYK-QIS 278
Query: 343 DVRFVPELKRNLISLSMFDGLGYCTRIEHGVCKI 376
+V + P+L ++L+S+ GY EH C I
Sbjct: 279 NVLYAPKLNQSLLSVPQLLTKGYKVLFEHEKCVI 380
>BF648150 similar to GP|14586969|gb| pol polyprotein {Citrus x paradisi},
partial (3%)
Length = 658
Score = 48.9 bits (115), Expect = 1e-05
Identities = 41/152 (26%), Positives = 70/152 (45%), Gaps = 9/152 (5%)
Frame = +2
Query: 42 AMPATLTQEEKREMIDKAK--SAIVLCLG-------DKVLRDVAREATAASM*AKLESLY 92
A P E E D+ K + +CLG D + +A + KLE+ Y
Sbjct: 122 ARPEDKDDETVAETRDRQKWDNDDYICLGHILNGMSDSLFDIYQSSPSAKDLWDKLETRY 301
Query: 93 MTKSLAHRQLLKQQLYSFKMVESISISEQLTEFNKILVDLANIEVNTEDEDKALLLLCSL 152
M + ++ L ++KMV++ S+ EQL E +IL + +N ++ ++ L
Sbjct: 302 MREDATSKKFLVSHFNNYKMVDNKSVMEQLYEIERILNNYKQHNMNMDETIIVSSIIDKL 481
Query: 153 PKSFEHFKDTILYGKEGTTTLEEVQAALRTKE 184
P S++ FK T+ + KE +LE++ LR E
Sbjct: 482 PPSWKDFKRTMKHKKE-DISLEQLGNHLRLXE 574
>AW736233 GP|21105451|gb small nuclear ribonucleoprotein D1 {Danio rerio},
partial (14%)
Length = 510
Score = 47.4 bits (111), Expect = 4e-05
Identities = 47/177 (26%), Positives = 77/177 (42%), Gaps = 3/177 (1%)
Frame = +1
Query: 122 LTEFNK-ILVDLANIEVNTEDEDKALLLLCSLPKSFEHFKDTILYGKEGTTTLEEVQAAL 180
L E+N +LV ++ + +E +++L++ FE ++ + T V A L
Sbjct: 7 LEEYNSFVLVIYIRLDSPSMEEGESILMMQEA--QFEKYRQEL--------TNPSVSANL 156
Query: 181 RTKEL-TKFKDLKVDE-GSEGLNVARGRNEHRGKGKGKSRSKSRSKGFDKSKYKCFLCHK 238
EL +K + + E G+E N RGR RG+G+G+ RS S ++ +C +C +
Sbjct: 157 AQSELPSKPGNSESQEVGTEYYNAGRGRGRGRGRGRGRGRSNS-------NRLQCQICAR 315
Query: 239 QGHFKKDCPDKGGDGSPSVQVAEASNEEGYESTGALVVTSWKSEKSWVLDSGCSYHM 295
H C + S S A E +S SE W DSG S+H+
Sbjct: 316 NNHDAARCCFRYDQASSSQAHHRAPPSE-------YAASSSYSEAPWYPDSGASHHL 465
>TC93065
Length = 783
Score = 44.3 bits (103), Expect = 3e-04
Identities = 39/176 (22%), Positives = 79/176 (44%), Gaps = 12/176 (6%)
Frame = +2
Query: 103 LKQQLYSFKMVESISISEQLTEFNKILVDLANIEVNTEDEDKALLLLCSLPKSFEHFKDT 162
L+++ + KM E+ ++ E +K++ + + + D+ +L LP+ FE +
Sbjct: 215 LRREFEALKMKETETVREFSDRISKVVTQIRLLGEDLSDQRVVEKILVCLPEMFEAKISS 394
Query: 163 ILYGKE-GTTTLEEVQAALRTKELTKFKDLKVDEGSEGLNVARGRNEHRG-KGKGKSRSK 220
+ K T+ E+ AL+ E + + L+++E EG +A + +++ K GK +
Sbjct: 395 LEENKNFSEITVAELVNALQASE--QRRSLRMEENVEGAFLANNKGKNQSFKSFGKKKFP 568
Query: 221 -----SRSKGFDKSKY-----KCFLCHKQGHFKKDCPDKGGDGSPSVQVAEASNEE 266
+ DK + KC C++ GH +K C +K +V E E+
Sbjct: 569 PCPHCKKDTHLDKFCWYRPGVKCRACNQLGHVEKVCKNKTNQQEQEARVVEHHQED 736
>AW773859
Length = 538
Score = 43.9 bits (102), Expect = 4e-04
Identities = 22/71 (30%), Positives = 35/71 (48%)
Frame = -3
Query: 417 LWHLRLGHVSERGLVELAKQGLLGKDKLDKLEFCEHCILGKQHRVKFGSGMHHSSRLFEY 476
LWH RLGH+S R L+ L +D+ C+ C + ++ F + +S+ +E
Sbjct: 230 LWHFRLGHLSNRKLLSLHSN--FPFITIDQNSVCDICHYSRHKKLPFQLSTNRASKCYEL 57
Query: 477 VHSDLLGPSKT 487
H D+ GP T
Sbjct: 56 FHFDIWGPFST 24
>BQ122739
Length = 575
Score = 38.1 bits (87), Expect = 0.022
Identities = 26/85 (30%), Positives = 43/85 (50%), Gaps = 4/85 (4%)
Frame = +1
Query: 359 MFDGLGYCTRIEHGVCKISHGALITVKGSKMNGLYILDGSIVIGNASV----ASVVPHNN 414
+ D GY IE G +I++ +++ +KG NGL +L G + A ++V +
Sbjct: 220 LLDDQGYIFNIEDGDLRITNDSMVLMKGKLENGLSLL*GRTSMDTADAIYIRCNLVSSRS 399
Query: 415 SELWHLRLGHVSERGLVELAKQGLL 439
S R+GHVS+ G+ +L G L
Sbjct: 400 SAY---RMGHVSKGGIKKLNTIGAL 465
>TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana, partial (91%)
Length = 860
Score = 37.0 bits (84), Expect = 0.050
Identities = 20/63 (31%), Positives = 29/63 (45%), Gaps = 3/63 (4%)
Frame = +3
Query: 195 EGSEGLNVARGRNEHRGKGKGKS---RSKSRSKGFDKSKYKCFLCHKQGHFKKDCPDKGG 251
+G G V N G G G R + G S KC+ C + GHF ++C ++GG
Sbjct: 216 DGKNGWRVELSHNSRSGGGGGGGGGGRGRGGGGG-GGSDLKCYECGEPGHFARECRNRGG 392
Query: 252 DGS 254
G+
Sbjct: 393 GGA 401
>TC87639 GP|9663153|emb|CAC01132.1 transport-secretion protein 2.2 (TTS-2.2)
{Homo sapiens}, partial (2%)
Length = 1522
Score = 37.0 bits (84), Expect = 0.050
Identities = 42/201 (20%), Positives = 82/201 (39%), Gaps = 19/201 (9%)
Frame = -1
Query: 86 AKLESLYMTKSLAHRQLLKQQLYSFKMVESISISEQLTEFNKILVD-----------LAN 134
A L+ Y+ ++ R + QL S + ++ ++ L F K+L D ++
Sbjct: 778 AYLDRTYLDPNIQSRAVA--QLQSLRQKDTERLATFLPRFEKVLADAGGYSWPDVVQISL 605
Query: 135 IEVNTEDEDKALLLLCSLPKSFEHFKDTILYGKEGTTTLEEVQAA-LRTKELTKFKDLKV 193
+E K LL+ LP + + + ++ +E ++ R T+ L V
Sbjct: 604 LETALVPRLKELLITVELPTVYSQWLSKV---QDIAWKMERMKTPPTRWAPATR---LPV 443
Query: 194 DEGSEGLNVARGRNEHRGKGKGKSRSKSRSKGFD---KSKYKCFLCHKQGHFKKDCPD-- 248
+ +G + G + + +G S S S ++G + +C+ CH++GH ++C +
Sbjct: 442 SKDRDGDMMMTGAIHKQRRRRGSSSSVSSAEGAPPPRRDMRECYSCHERGHIARNCTNTS 263
Query: 249 --KGGDGSPSVQVAEASNEEG 267
K P V E EG
Sbjct: 262 AAKKKKKGPKVAKVEPQKAEG 200
>TC84935 similar to PIR|G96631|G96631 probable RNA-binding protein F8A5.17
[imported] - Arabidopsis thaliana, partial (41%)
Length = 552
Score = 34.7 bits (78), Expect = 0.25
Identities = 12/21 (57%), Positives = 15/21 (71%)
Frame = +2
Query: 233 CFLCHKQGHFKKDCPDKGGDG 253
CF C + GH+ +DCP GGDG
Sbjct: 473 CFKCGRPGHWARDCPLAGGDG 535
>TC83030 weakly similar to GP|18855061|gb|AAL79753.1 putative RNA helicase
{Oryza sativa}, partial (7%)
Length = 624
Score = 33.9 bits (76), Expect = 0.42
Identities = 15/38 (39%), Positives = 19/38 (49%)
Frame = +1
Query: 215 GKSRSKSRSKGFDKSKYKCFLCHKQGHFKKDCPDKGGD 252
G+ S+S S CF C + GH DCP+K GD
Sbjct: 112 GRQSSRSSSSPNRSFAGTCFTCGESGHRASDCPNKRGD 225
Score = 30.4 bits (67), Expect = 4.7
Identities = 14/32 (43%), Positives = 17/32 (52%), Gaps = 1/32 (3%)
Frame = +1
Query: 1140 EWVFEGRFEVHKSSSRERRFGGVC-*CGQCGH 1170
+W+ GR SSS R F G C CG+ GH
Sbjct: 97 DWLIGGRQSSRSSSSPNRSFAGTCFTCGESGH 192
>TC83119 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (16%)
Length = 421
Score = 33.9 bits (76), Expect = 0.42
Identities = 22/79 (27%), Positives = 37/79 (45%), Gaps = 13/79 (16%)
Frame = +3
Query: 183 KELTKFKDLKVDEGSEGLNVARGRNEHRGKGKG--KSRS-----------KSRSKGFDKS 229
KE+ K K+LK+ S + +R R+ R + K RS + S+GF +
Sbjct: 162 KEVKKEKNLKMSSDSRSRSRSRSRSRSRSRSPRIRKIRSDRHSYRDAPYRRDSSRGFSRD 341
Query: 230 KYKCFLCHKQGHFKKDCPD 248
C C + GH+ ++CP+
Sbjct: 342 NL-CKNCKRPGHYARECPN 395
>BG450974 similar to PIR|T05112|T05 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana, partial (54%)
Length = 364
Score = 33.5 bits (75), Expect = 0.55
Identities = 16/52 (30%), Positives = 23/52 (43%)
Frame = +1
Query: 195 EGSEGLNVARGRNEHRGKGKGKSRSKSRSKGFDKSKYKCFLCHKQGHFKKDC 246
+G G V N G G G+ + G D KC+ C + GHF ++C
Sbjct: 214 DGKNGWRVQLSHNSKSGGGGGRGGGRGGRGGDD---LKCYECGEPGHFAREC 360
>AJ388976 similar to PIR|E84638|E84 probable RSZp22 splicing factor
[imported] - Arabidopsis thaliana, partial (62%)
Length = 508
Score = 32.7 bits (73), Expect = 0.94
Identities = 18/45 (40%), Positives = 22/45 (48%), Gaps = 1/45 (2%)
Frame = +2
Query: 210 RGKGKGKSRSKSRSKGFDKSKYKCFLCHKQGHFKKDC-PDKGGDG 253
RG G G + RS G S KC+ C + GHF + C GG G
Sbjct: 278 RGGGGGGGGGRGRSGG-GGSDLKCYXCGEPGHFARXCNSSPGGSG 409
>TC79595 similar to GP|3850823|emb|CAA77136.1 U2 snRNP auxiliary factor
large subunit {Nicotiana plumbaginifolia}, partial (77%)
Length = 1525
Score = 31.6 bits (70), Expect = 2.1
Identities = 14/26 (53%), Positives = 20/26 (76%), Gaps = 1/26 (3%)
Frame = +2
Query: 203 ARGRNEHRGKGKGKSRSKS-RSKGFD 227
+R R+EHR + + +SRSKS R+ GFD
Sbjct: 89 SRARSEHRSRSRSRSRSKSKRTSGFD 166
>TC81417 weakly similar to GP|10998142|dbj|BAB03113.
gene_id:MEC18.14~ref|NP_037458.1~similar to unknown
protein {Arabidopsis thaliana}, partial (23%)
Length = 841
Score = 31.2 bits (69), Expect = 2.7
Identities = 11/34 (32%), Positives = 22/34 (64%)
Frame = +1
Query: 392 LYILDGSIVIGNASVASVVPHNNSELWHLRLGHV 425
+Y+L + +GN S ++ P + ++LW LR+ H+
Sbjct: 10 MYLL*KEMQLGNRSYTNLPPESTAKLWTLRMNHI 111
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.346 0.154 0.537
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 42,518,187
Number of Sequences: 36976
Number of extensions: 642691
Number of successful extensions: 4857
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 1993
Number of HSP's successfully gapped in prelim test: 209
Number of HSP's that attempted gapping in prelim test: 2684
Number of HSP's gapped (non-prelim): 2423
length of query: 1307
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1199
effective length of database: 5,021,319
effective search space: 6020561481
effective search space used: 6020561481
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC124217.5


