

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124215.2 + phase: 0
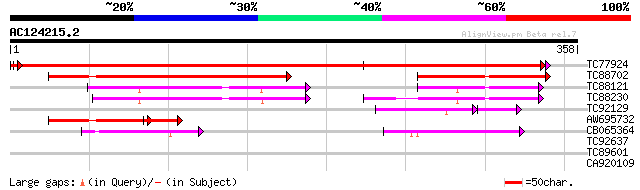
(358 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77924 similar to PIR|H84807|H84807 probable phospholipid cytid... 672 0.0
TC88702 similar to GP|13129435|gb|AAK13093.1 Putative phospholip... 220 7e-58
TC88121 homologue to PIR|T06558|T06558 choline-phosphate cytidyl... 110 1e-24
TC88230 similar to PIR|T07981|T07981 probable choline-phosphate ... 104 6e-23
TC92129 homologue to PIR|H84807|H84807 probable phospholipid cyt... 90 4e-20
AW695732 similar to GP|13129435|gb| Putative phospholipid cytidy... 73 5e-16
CB065364 homologue to PIR|A83022|A83 LPS biosynthesis protein Rf... 58 6e-09
TC92637 similar to PIR|T39903|T39903 serine-rich protein - fissi... 29 3.2
TC89601 similar to GP|9652289|gb|AAF91469.1| putative vacuolar p... 28 5.5
CA920109 similar to GP|22535636|dbj hypothetical protein~predict... 27 9.3
>TC77924 similar to PIR|H84807|H84807 probable phospholipid
cytidylyltransferase [imported] - Arabidopsis thaliana,
partial (86%)
Length = 1691
Score = 672 bits (1733), Expect(2) = 0.0
Identities = 330/336 (98%), Positives = 332/336 (98%)
Frame = +2
Query: 3 KMGGITSSVESYAAVKWVVGVIIVGVSVLGFYSSSGPFGLWGKNGRRRNKKPIRVYMDGC 62
K G SSVESYAAVKWVVGVIIVGVSVLGFYSSSGPFGLWGKNGRRRNKKPIRVYMDGC
Sbjct: 74 KWGVSPSSVESYAAVKWVVGVIIVGVSVLGFYSSSGPFGLWGKNGRRRNKKPIRVYMDGC 253
Query: 63 FDMMHYGHCNALRQARALGDQLIVGVVSDDEIIANKGPPVTPLHERLIMVNAVKWVDEVI 122
FDMMHYGHCNALRQARALGDQLIVGVVSDDEIIANKGPPVTPLHERLIMVNAVKWVDEVI
Sbjct: 254 FDMMHYGHCNALRQARALGDQLIVGVVSDDEIIANKGPPVTPLHERLIMVNAVKWVDEVI 433
Query: 123 PEAPYAITEEFMKKLFDEYNIDYIIHGDDPCVLPDGTDAYAHAKKAGRYKQIKRTEGVSS 182
PEAPYAITEEFMKKLFDEYNIDYIIHGDDPCVLPDGTDAYAHAKKAGRYKQIKRTEGVSS
Sbjct: 434 PEAPYAITEEFMKKLFDEYNIDYIIHGDDPCVLPDGTDAYAHAKKAGRYKQIKRTEGVSS 613
Query: 183 TDIVGRMLLCVRERNIADTHNHSSLQRQFSNGRGQQKFEDGGVVASGTRVSHFLPTSRRI 242
TDIVGRMLLCVRERNIADTHNHSSLQRQFSNGRGQQKFEDGGVVASGTRVSHFLPTSRRI
Sbjct: 614 TDIVGRMLLCVRERNIADTHNHSSLQRQFSNGRGQQKFEDGGVVASGTRVSHFLPTSRRI 793
Query: 243 VQFSNGRSPGPDARIVYIDGAFDLFHAGHVEILRLARDRGDFLLVGIHTDQTVSATRGLH 302
VQFSNGRSPGPDARIVYIDGAFDLFHAGHVEILRLARDRGDFLLVGIHTDQTVSATRGLH
Sbjct: 794 VQFSNGRSPGPDARIVYIDGAFDLFHAGHVEILRLARDRGDFLLVGIHTDQTVSATRGLH 973
Query: 303 RPIMSLHERSLSVLACRYVDEVIIGAPWEVSKDMVS 338
RPIMSLHERSLSVLACRYVDEVIIGAPWEVSKDM++
Sbjct: 974 RPIMSLHERSLSVLACRYVDEVIIGAPWEVSKDMIT 1081
Score = 76.6 bits (187), Expect = 1e-14
Identities = 45/119 (37%), Positives = 66/119 (54%), Gaps = 1/119 (0%)
Frame = +2
Query: 224 GVVASGTRVSHFLPTSRRIVQFS-NGRSPGPDARIVYIDGAFDLFHAGHVEILRLARDRG 282
GV+ G V F +S + NGR VY+DG FD+ H GH LR AR G
Sbjct: 131 GVIIVGVSVLGFYSSSGPFGLWGKNGRRRNKKPIRVYMDGCFDMMHYGHCNALRQARALG 310
Query: 283 DFLLVGIHTDQTVSATRGLHRPIMSLHERSLSVLACRYVDEVIIGAPWEVSKDMVSLIF 341
D L+VG+ +D + A +G P+ LHER + V A ++VDEVI AP+ ++++ + +F
Sbjct: 311 DQLIVGVVSDDEIIANKG--PPVTPLHERLIMVNAVKWVDEVIPEAPYAITEEFMKKLF 481
Score = 20.8 bits (42), Expect(2) = 0.0
Identities = 8/8 (100%), Positives = 8/8 (100%)
Frame = +1
Query: 1 MNKMGGIT 8
MNKMGGIT
Sbjct: 67 MNKMGGIT 90
>TC88702 similar to GP|13129435|gb|AAK13093.1 Putative phospholipid
cytidylyltransferase {Oryza sativa}, partial (17%)
Length = 801
Score = 220 bits (560), Expect = 7e-58
Identities = 105/154 (68%), Positives = 125/154 (80%)
Frame = +1
Query: 25 IVGVSVLGFYSSSGPFGLWGKNGRRRNKKPIRVYMDGCFDMMHYGHCNALRQARALGDQL 84
++GV L S G K G+RR +RVYMDGCFD+MHYGH NALRQA+ALGD+L
Sbjct: 352 VIGVPSLPLPCSWYNLGKKKKTGKRR----VRVYMDGCFDLMHYGHANALRQAKALGDEL 519
Query: 85 IVGVVSDDEIIANKGPPVTPLHERLIMVNAVKWVDEVIPEAPYAITEEFMKKLFDEYNID 144
+VG+VSD+EI+ANKGPPV + ERL +V+ +KWVDEVI +APYAITE F+ +LF EYNID
Sbjct: 520 VVGLVSDEEIVANKGPPVLSMDERLALVSGLKWVDEVITDAPYAITETFLNRLFHEYNID 699
Query: 145 YIIHGDDPCVLPDGTDAYAHAKKAGRYKQIKRTE 178
Y+IHGDDPC+LPDGTDAYA AKKAGRYKQIKRTE
Sbjct: 700 YVIHGDDPCLLPDGTDAYAAAKKAGRYKQIKRTE 801
Score = 69.7 bits (169), Expect = 2e-12
Identities = 34/84 (40%), Positives = 54/84 (63%)
Frame = +1
Query: 258 VYIDGAFDLFHAGHVEILRLARDRGDFLLVGIHTDQTVSATRGLHRPIMSLHERSLSVLA 317
VY+DG FDL H GH LR A+ GD L+VG+ +D+ + A +G P++S+ ER V
Sbjct: 436 VYMDGCFDLMHYGHANALRQAKALGDELVVGLVSDEEIVANKG--PPVLSMDERLALVSG 609
Query: 318 CRYVDEVIIGAPWEVSKDMVSLIF 341
++VDEVI AP+ +++ ++ +F
Sbjct: 610 LKWVDEVITDAPYAITETFLNRLF 681
>TC88121 homologue to PIR|T06558|T06558 choline-phosphate
cytidylyltransferase (EC 2.7.7.15) - garden pea, partial
(97%)
Length = 1063
Score = 110 bits (274), Expect = 1e-24
Identities = 58/145 (40%), Positives = 84/145 (57%), Gaps = 4/145 (2%)
Frame = +3
Query: 50 RNKKPIRVYMDGCFDMMHYGHCNALRQARAL--GDQLIVGVVSDDEIIANKGPPVTPLHE 107
+N KP+RVY DG +D+ H+GH +L QA+ L+VG SD+ KG V E
Sbjct: 126 KNDKPVRVYADGIYDLFHFGHARSLEQAKKSFPNTYLLVGCCSDEITHKYKGKTVMNEQE 305
Query: 108 RLIMVNAVKWVDEVIPEAPYAITEEFMKKLFDEYNIDYIIHGDDPCVLPD--GTDAYAHA 165
R + KWVDEVIP+ P+ I +EF+ D++ IDY+ H P G D Y
Sbjct: 306 RYESLRHCKWVDEVIPDVPWVINQEFI----DKHKIDYVAHDALPYADTSGAGNDVYEFV 473
Query: 166 KKAGRYKQIKRTEGVSSTDIVGRML 190
K G++K+ KRTEG+S++DI+ R++
Sbjct: 474 KAIGKFKETKRTEGISTSDIIMRII 548
Score = 57.4 bits (137), Expect = 8e-09
Identities = 30/82 (36%), Positives = 47/82 (56%), Gaps = 2/82 (2%)
Frame = +3
Query: 258 VYIDGAFDLFHAGHVEILRLARDR--GDFLLVGIHTDQTVSATRGLHRPIMSLHERSLSV 315
VY DG +DLFH GH L A+ +LLVG +D+ +G + +M+ ER S+
Sbjct: 147 VYADGIYDLFHFGHARSLEQAKKSFPNTYLLVGCCSDEITHKYKG--KTVMNEQERYESL 320
Query: 316 LACRYVDEVIIGAPWEVSKDMV 337
C++VDEVI PW ++++ +
Sbjct: 321 RHCKWVDEVIPDVPWVINQEFI 386
>TC88230 similar to PIR|T07981|T07981 probable choline-phosphate
cytidylyltransferase (EC 2.7.7.15) (clone CCT1) - rape,
partial (59%)
Length = 1364
Score = 104 bits (259), Expect = 6e-23
Identities = 56/142 (39%), Positives = 81/142 (56%), Gaps = 4/142 (2%)
Frame = +3
Query: 53 KPIRVYMDGCFDMMHYGHCNALRQARAL--GDQLIVGVVSDDEIIANKGPPVTPLHERLI 110
+P+RVY DG +D+ H+GH +L QA+ L+VG SD KG V ER
Sbjct: 114 RPVRVYADGIYDLFHFGHARSLEQAKKSFPNTYLLVGCCSDAVTHKYKGKTVMTEDERYE 293
Query: 111 MVNAVKWVDEVIPEAPYAITEEFMKKLFDEYNIDYIIHGDDPCVLPDG--TDAYAHAKKA 168
+ KWVDEVIP+AP+ I +EF+ D+ ID++ H P G D Y K
Sbjct: 294 SLRHCKWVDEVIPDAPWVINQEFL----DKNKIDFVAHDSLPYADTSGAANDVYEFVKAV 461
Query: 169 GRYKQIKRTEGVSSTDIVGRML 190
GR+K+ +RTEG+S++DI+ R++
Sbjct: 462 GRFKETQRTEGISTSDIIMRIV 527
Score = 57.8 bits (138), Expect = 6e-09
Identities = 37/116 (31%), Positives = 57/116 (48%), Gaps = 2/116 (1%)
Frame = +3
Query: 224 GVVASGTRVSHFLPTSRRIVQFSNGRSPGPDARIVYIDGAFDLFHAGHVEILRLARDR-- 281
G + S + + H P R V+ VY DG +DLFH GH L A+
Sbjct: 63 GNMTSSSTIPHLPPPEDRPVR-------------VYADGIYDLFHFGHARSLEQAKKSFP 203
Query: 282 GDFLLVGIHTDQTVSATRGLHRPIMSLHERSLSVLACRYVDEVIIGAPWEVSKDMV 337
+LLVG +D +G + +M+ ER S+ C++VDEVI APW ++++ +
Sbjct: 204 NTYLLVGCCSDAVTHKYKG--KTVMTEDERYESLRHCKWVDEVIPDAPWVINQEFL 365
>TC92129 homologue to PIR|H84807|H84807 probable phospholipid
cytidylyltransferase [imported] - Arabidopsis thaliana,
partial (14%)
Length = 510
Score = 89.7 bits (221), Expect(2) = 4e-20
Identities = 51/113 (45%), Positives = 60/113 (52%), Gaps = 49/113 (43%)
Frame = +1
Query: 232 VSHFLPTSRRIVQFSNGRSPGPDARIVYIDGAFDLFHAGHVEI----------------- 274
+S FLPTSRRIVQFSNG+ PGP+ARIVYIDGAFDLFHAGHV++
Sbjct: 4 ISQFLPTSRRIVQFSNGKGPGPNARIVYIDGAFDLFHAGHVQV*IGFM*GVVYYPQLNA* 183
Query: 275 --------------------------------LRLARDRGDFLLVGIHTDQTV 295
L+ AR+ GDFLLVGIH+D+TV
Sbjct: 184 VHGSP*PNRQEPFDPIILI*LCLIHYFVLYQMLKRARELGDFLLVGIHSDETV 342
Score = 27.7 bits (60), Expect = 7.1
Identities = 11/25 (44%), Positives = 14/25 (56%)
Frame = +1
Query: 46 NGRRRNKKPIRVYMDGCFDMMHYGH 70
NG+ VY+DG FD+ H GH
Sbjct: 49 NGKGPGPNARIVYIDGAFDLFHAGH 123
Score = 25.8 bits (55), Expect(2) = 4e-20
Identities = 16/29 (55%), Positives = 16/29 (55%), Gaps = 1/29 (3%)
Frame = +2
Query: 296 SATRGLHRPIMSLHERSL-SVLACRYVDE 323
S RG H PIM LHE A RYVDE
Sbjct: 419 SENRGNHYPIMHLHEAEP*RG*ASRYVDE 505
>AW695732 similar to GP|13129435|gb| Putative phospholipid
cytidylyltransferase {Oryza sativa}, partial (7%)
Length = 497
Score = 72.8 bits (177), Expect(2) = 5e-16
Identities = 36/66 (54%), Positives = 46/66 (69%)
Frame = +3
Query: 25 IVGVSVLGFYSSSGPFGLWGKNGRRRNKKPIRVYMDGCFDMMHYGHCNALRQARALGDQL 84
++GV L S G K G+RR + VYMDGCFD+MHYGH NAL+QA+ALGD+L
Sbjct: 252 VIGVPSLPLPCSWYNLGKKKKTGKRR----VHVYMDGCFDLMHYGHANALKQAKALGDEL 419
Query: 85 IVGVVS 90
+VG+VS
Sbjct: 420 VVGLVS 437
Score = 39.7 bits (91), Expect = 0.002
Identities = 17/32 (53%), Positives = 22/32 (68%)
Frame = +3
Query: 258 VYIDGAFDLFHAGHVEILRLARDRGDFLLVGI 289
VY+DG FDL H GH L+ A+ GD L+VG+
Sbjct: 336 VYMDGCFDLMHYGHANALKQAKALGDELVVGL 431
Score = 28.9 bits (63), Expect(2) = 5e-16
Identities = 10/25 (40%), Positives = 19/25 (76%)
Frame = +1
Query: 85 IVGVVSDDEIIANKGPPVTPLHERL 109
++ ++ D+EI+ANKGPPV + ++
Sbjct: 421 LLDLLVDEEIVANKGPPVLSMDRKV 495
>CB065364 homologue to PIR|A83022|A83 LPS biosynthesis protein RfaE PA4996
[imported] - Pseudomonas aeruginosa (strain PAO1),
partial (47%)
Length = 682
Score = 57.8 bits (138), Expect = 6e-09
Identities = 39/105 (37%), Positives = 51/105 (48%), Gaps = 16/105 (15%)
Frame = +2
Query: 237 PTSRRIVQFSNGRSPG-----------PDAR-----IVYIDGAFDLFHAGHVEILRLARD 280
P RR +Q G G DAR IV+ +G FD+ HAGHV L AR
Sbjct: 341 PELRRAIQREEGSERGVLGLEQLLLAIDDARAHNEKIVFTNGCFDILHAGHVTYLEQARA 520
Query: 281 RGDFLLVGIHTDQTVSATRGLHRPIMSLHERSLSVLACRYVDEVI 325
+GD L+V ++ D +VS +G RPI S+ R + VD VI
Sbjct: 521 QGDRLIVAVNDDASVSRLKGPGRPINSVDRRMAVLAGLGAVDWVI 655
Score = 55.8 bits (133), Expect = 2e-08
Identities = 32/79 (40%), Positives = 46/79 (57%), Gaps = 2/79 (2%)
Frame = +2
Query: 46 NGRRRNKKPIRVYMDGCFDMMHYGHCNALRQARALGDQLIVGVVSDDEIIANKGP--PVT 103
+ R N+K V+ +GCFD++H GH L QARA GD+LIV V D + KGP P+
Sbjct: 425 DARAHNEKI--VFTNGCFDILHAGHVTYLEQARAQGDRLIVAVNDDASVSRLKGPGRPIN 598
Query: 104 PLHERLIMVNAVKWVDEVI 122
+ R+ ++ + VD VI
Sbjct: 599 SVDRRMAVLAGLGAVDWVI 655
>TC92637 similar to PIR|T39903|T39903 serine-rich protein - fission yeast
(Schizosaccharomyces pombe), partial (7%)
Length = 671
Score = 28.9 bits (63), Expect = 3.2
Identities = 19/50 (38%), Positives = 27/50 (54%)
Frame = +3
Query: 201 THNHSSLQRQFSNGRGQQKFEDGGVVASGTRVSHFLPTSRRIVQFSNGRS 250
+H+HSS+Q QFS QKF+ + RVS + RI+ FS +S
Sbjct: 81 SHSHSSIQFQFS-----QKFQ*S*PIFGSARVSFPYKRTPRIIVFSFTKS 215
>TC89601 similar to GP|9652289|gb|AAF91469.1| putative vacuolar proton
ATPase subunit E {Lycopersicon esculentum}, partial
(52%)
Length = 832
Score = 28.1 bits (61), Expect = 5.5
Identities = 18/63 (28%), Positives = 31/63 (48%), Gaps = 2/63 (3%)
Frame = -3
Query: 295 VSATRGLHRPIMSLHERSLSVLACR-YVDEVIIGAPWEVSKDMVS-LIFACLFFMLFVYD 352
+S TR H P ++ S+ +L CR +D V+ W ++ S L+ +C+ M +
Sbjct: 368 LSITRSQHHPTRAVRFVSIVMLGCRGKIDTVVNNDLWWINTCFFSILLCSCIQHMFYQVH 189
Query: 353 VSF 355
V F
Sbjct: 188 VMF 180
>CA920109 similar to GP|22535636|dbj hypothetical protein~predicted by
GENSCAN etc. {Oryza sativa (japonica cultivar-group)},
partial (12%)
Length = 625
Score = 27.3 bits (59), Expect = 9.3
Identities = 14/39 (35%), Positives = 21/39 (52%), Gaps = 1/39 (2%)
Frame = -2
Query: 148 HGDDPCVLPD-GTDAYAHAKKAGRYKQIKRTEGVSSTDI 185
H DPC +PD G + KK G+ K+ K + ST++
Sbjct: 363 HLVDPCTVPDVGDIKKSKKKKKGKEKETKSSSNGDSTEL 247
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.324 0.141 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,023,172
Number of Sequences: 36976
Number of extensions: 150645
Number of successful extensions: 742
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 719
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 732
length of query: 358
length of database: 9,014,727
effective HSP length: 97
effective length of query: 261
effective length of database: 5,428,055
effective search space: 1416722355
effective search space used: 1416722355
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC124215.2


