

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123975.9 + phase: 0 /pseudo
(622 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
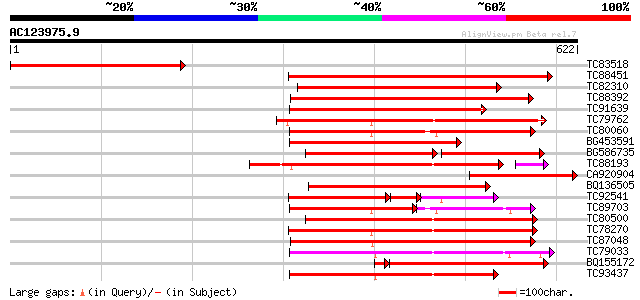
Score E
Sequences producing significant alignments: (bits) Value
TC83518 391 e-109
TC88451 similar to GP|836954|gb|AAC23542.1|| receptor protein ki... 337 1e-92
TC82310 similar to PIR|T05181|T05181 S-receptor kinase (EC 2.7.1... 304 6e-83
TC88392 similar to GP|11275527|dbj|BAB18292. putative receptor-l... 285 5e-77
TC91639 similar to PIR|T05181|T05181 S-receptor kinase (EC 2.7.1... 284 6e-77
TC79762 similar to PIR|H96731|H96731 hypothetical protein F5A18.... 281 7e-76
TC80060 similar to PIR|D96574|D96574 hypothetical protein T3F20.... 264 7e-71
BG453591 similar to PIR|T05181|T05 S-receptor kinase (EC 2.7.1.-... 260 1e-69
BG586735 similar to GP|11275527|db putative receptor-like protei... 180 2e-67
TC88193 similar to PIR|G96602|G96602 probable receptor protein k... 244 1e-66
CA920904 similar to GP|22208478|gb| receptor-like kinase {Sorghu... 246 1e-65
BQ136505 similar to PIR|T05754|T05 S-receptor kinase (EC 2.7.1.-... 244 5e-65
TC92541 similar to PIR|T02153|T02153 protein kinase homolog T1F1... 135 6e-64
TC89703 similar to GP|11994595|dbj|BAB02650. receptor-like serin... 172 5e-63
TC80500 similar to PIR|G96602|G96602 probable receptor protein k... 230 1e-60
TC78270 similar to GP|16649103|gb|AAL24403.1 Unknown protein {Ar... 223 1e-58
TC87048 similar to GP|8778594|gb|AAF79602.1| F5M15.3 {Arabidopsi... 222 4e-58
TC79033 similar to GP|21554229|gb|AAM63304.1 somatic embryogenes... 215 4e-56
BQ155172 weakly similar to GP|6686398|gb F1E22.15 {Arabidopsis t... 208 3e-55
TC93437 similar to GP|11227578|emb|CAC16506. unnamed protein pro... 211 7e-55
>TC83518
Length = 666
Score = 391 bits (1004), Expect = e-109
Identities = 192/193 (99%), Positives = 193/193 (99%)
Frame = +1
Query: 1 MFHYPIVFKFIQFFLVLYTLDFLSYLALADPPYEICSTRNIYANGSSFDNNLSNLLLSLP 60
MFHYPI+FKFIQFFLVLYTLDFLSYLALADPPYEICSTRNIYANGSSFDNNLSNLLLSLP
Sbjct: 88 MFHYPILFKFIQFFLVLYTLDFLSYLALADPPYEICSTRNIYANGSSFDNNLSNLLLSLP 267
Query: 61 FNDSNSISKFGNTSSGIGLDRVYGLYMCLDFVSNETCLKCVTNAIADTVKLCPQSKEAVV 120
FNDSNSISKFGNTSSGIGLDRVYGLYMCLDFVSNETCLKCVTNAIADTVKLCPQSKEAVV
Sbjct: 268 FNDSNSISKFGNTSSGIGLDRVYGLYMCLDFVSNETCLKCVTNAIADTVKLCPQSKEAVV 447
Query: 121 YEEFCQVRYSNKNFIGSLNVNGNIGKDNVQNISEPVKFETSVNKLLNGLTKIASFNVSAN 180
YEEFCQVRYSNKNFIGSLNVNGNIGKDNVQNISEPVKFETSVNKLLNGLTKIASFNVSAN
Sbjct: 448 YEEFCQVRYSNKNFIGSLNVNGNIGKDNVQNISEPVKFETSVNKLLNGLTKIASFNVSAN 627
Query: 181 MYATGEVPFEDKT 193
MYATGEVPFEDKT
Sbjct: 628 MYATGEVPFEDKT 666
>TC88451 similar to GP|836954|gb|AAC23542.1|| receptor protein kinase
{Ipomoea trifida}, partial (28%)
Length = 1276
Score = 337 bits (863), Expect = 1e-92
Identities = 171/294 (58%), Positives = 219/294 (74%), Gaps = 4/294 (1%)
Frame = +3
Query: 306 SVNFTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNL 365
++ F+ NKLG+GG+GPVYKGIL+ GQE+A+KRLS S QG EF NE++LI +LQHKNL
Sbjct: 219 TMEFSPENKLGQGGYGPVYKGILATGQEIAVKRLSKTSGQGIVEFKNELLLICELQHKNL 398
Query: 366 VKLLGFCVDGEEKLLVYEYLPNGSLDVVLFE--QHAQLDWTKRLDIINGIARGILYLHED 423
V+LLG C+ EE++L+YEY+PN SLD LF+ + LDW KR +II GIA+G+LYLH+
Sbjct: 399 VQLLGCCIHEEERILIYEYMPNKSLDFYLFDCTKKKLLDWKKRFNIIEGIAQGLLYLHKY 578
Query: 424 SRLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAME 483
SRL+IIHRDLKASN+LLD +MNPKI+DFGMAR+F E NT IVGTYGYM+PEYAME
Sbjct: 579 SRLKIIHRDLKASNILLDENMNPKIADFGMARMFTQQESVVNTNRIVGTYGYMSPEYAME 758
Query: 484 GLYSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAWHLWNDGKGLELRDPLL-- 541
G+ S KSDV+ FGVLLLEI+ GI+N F +L+ +AW LWNDG+ L+L DP L
Sbjct: 759 GVCSTKSDVYSFGVLLLEIVCGIKNNSFYDVDRPLNLIGHAWELWNDGEYLKLMDPTLND 938
Query: 542 LCPGDQFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMNESVMLGQPGKPPFSVGR 595
D+ R +++GLLCV++ A DRPTMS V+ +L N+ V+ P KP F V R
Sbjct: 939 TFVPDEVKRCIHVGLLCVEQYANDRPTMSEVISVLTNKYVLTNLPRKPAFYVRR 1100
>TC82310 similar to PIR|T05181|T05181 S-receptor kinase (EC 2.7.1.-)
T6K22.120 precursor - Arabidopsis thaliana, partial
(24%)
Length = 696
Score = 304 bits (779), Expect = 6e-83
Identities = 149/226 (65%), Positives = 183/226 (80%), Gaps = 2/226 (0%)
Frame = +1
Query: 316 GEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVKLLGFCVDG 375
G+GGFG VYKG+L+D +E+A+KRLS S QG EE NE++LI KLQH+NLV+LL C++
Sbjct: 1 GKGGFGTVYKGVLADEKEIAVKRLSKTSSQGVEELKNEIILIAKLQHRNLVRLLACCIEQ 180
Query: 376 EEKLLVYEYLPNGSLDVVLFE--QHAQLDWTKRLDIINGIARGILYLHEDSRLQIIHRDL 433
EKLL+YEYLPN SLD LF+ + AQL W +RL+IINGIA+G+LYLHEDSRL++IHRDL
Sbjct: 181 NEKLLIYEYLPNSSLDFHLFDMVKGAQLAWRQRLNIINGIAKGLLYLHEDSRLRVIHRDL 360
Query: 434 KASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEGLYSIKSDVF 493
KASN+LLD +MNPKISDFG+AR F G + EANT +VGTYGYMAPEYAMEGL+S+KSDVF
Sbjct: 361 KASNILLDQEMNPKISDFGLARTFGGDQDEANTIRVVGTYGYMAPEYAMEGLFSVKSDVF 540
Query: 494 GFGVLLLEIITGIRNAGFCYSKTTPSLLAYAWHLWNDGKGLELRDP 539
FGVLLLEII+G +N+ F S+ SL +AW+LW KG EL DP
Sbjct: 541 SFGVLLLEIISGRKNSKFYLSEHGQSLPIFAWNLWCKRKGFELMDP 678
>TC88392 similar to GP|11275527|dbj|BAB18292. putative receptor-like protein
kinase {Oryza sativa (japonica cultivar-group)}, partial
(61%)
Length = 1017
Score = 285 bits (728), Expect = 5e-77
Identities = 142/267 (53%), Positives = 189/267 (70%), Gaps = 1/267 (0%)
Frame = +1
Query: 309 FTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVKL 368
F++ N+LG GGFGPV+KG++ +G+EVAIK+LS+ S QG EF NEV L+L++QHKNLV L
Sbjct: 217 FSELNQLGRGGFGPVFKGLMPNGEEVAIKKLSMESRQGIREFTNEVRLLLRIQHKNLVTL 396
Query: 369 LGFCVDGEEKLLVYEYLPNGSLDVVLFEQHAQLDWTKRLDIINGIARGILYLHEDSRLQI 428
LG C +G EK+LVYEYLPN SLD LF++ LDW R I+ GIARG+LYLHE++ +I
Sbjct: 397 LGCCAEGPEKMLVYEYLPNKSLDHFLFDKKRSLDWMTRFRIVTGIARGLLYLHEEAPERI 576
Query: 429 IHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEGLYSI 488
IHRD+KASN+LLD +NPKISDFG+AR+F G + T I T+GYMAPEYA+ G S+
Sbjct: 577 IHRDIKASNILLDEKLNPKISDFGLARLFPGEDTHVQTFRISRTHGYMAPEYALRGYLSV 756
Query: 489 KSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAWHLWNDGKGLELRDPLL-LCPGDQ 547
K+DVF +GVL+LEI++G +N LL+YAW L+ K ++L D + GD+
Sbjct: 757 KTDVFSYGVLVLEIVSGRKNHDLKLDAEKADLLSYAWKLYQGRKIMDLIDQNIGKYNGDE 936
Query: 548 FLRYMNIGLLCVQEDAFDRPTMSSVVL 574
+ +GLLC Q +RP M+SV L
Sbjct: 937 AAMCIQLGLLCCQASLVERPDMNSVNL 1017
>TC91639 similar to PIR|T05181|T05181 S-receptor kinase (EC 2.7.1.-)
T6K22.120 precursor - Arabidopsis thaliana, partial
(28%)
Length = 1313
Score = 284 bits (727), Expect = 6e-77
Identities = 140/218 (64%), Positives = 176/218 (80%), Gaps = 2/218 (0%)
Frame = +3
Query: 308 NFTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVK 367
NF++ NKLG+GGFGPVYKG L G+E+A+KRLS S QG +EF NE+ L +LQH+NLVK
Sbjct: 405 NFSEENKLGQGGFGPVYKGKLPSGEEIAVKRLSRRSGQGLDEFKNEMRLFAQLQHRNLVK 584
Query: 368 LLGFCVDGEEKLLVYEYLPNGSLDVVLFE--QHAQLDWTKRLDIINGIARGILYLHEDSR 425
L+G ++G+EKLLVYE++ N SLD LF+ + QLDW +R +II GIARG+LYLH DSR
Sbjct: 585 LMGCSIEGDEKLLVYEFMLNKSLDRFLFDPIKKTQLDWARRYEIIEGIARGLLYLHRDSR 764
Query: 426 LQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEGL 485
L+IIHRDLKASN+LLD +MNPKISDFG+ARIF G++ E N T +VGTYGYM+PEYAMEGL
Sbjct: 765 LRIIHRDLKASNILLDENMNPKISDFGLARIFGGNQNEENATKVVGTYGYMSPEYAMEGL 944
Query: 486 YSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAY 523
S+KSDV+ FGVLLLEI++G RN F +S + SL+ Y
Sbjct: 945 VSVKSDVYSFGVLLLEIVSGRRNTSFRHSDDS-SLIGY 1055
>TC79762 similar to PIR|H96731|H96731 hypothetical protein F5A18.8
[imported] - Arabidopsis thaliana, partial (55%)
Length = 1564
Score = 281 bits (718), Expect = 7e-76
Identities = 151/306 (49%), Positives = 204/306 (66%), Gaps = 9/306 (2%)
Frame = +3
Query: 293 LRNRQSKLFGD----CISVNFTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSE 348
+ +R+ K+F + NF ++KLGEGGFGPVYKG LSDG+EVA+K+LS S QG +
Sbjct: 324 MASREQKIFSYETLLSATKNFNATHKLGEGGFGPVYKGKLSDGREVAVKKLSQTSNQGKK 503
Query: 349 EFINEVMLILKLQHKNLVKLLGFCVDGEEKLLVYEYLPNGSLDVVLF---EQHAQLDWTK 405
EF+NE L+ ++QHKN+V LLG+CV G EK+LVYEY+P+ SLD LF E+ QLDW +
Sbjct: 504 EFMNEAKLLARVQHKNVVNLLGYCVHGTEKILVYEYVPHESLDKFLFKEAEKREQLDWKR 683
Query: 406 RLDIINGIARGILYLHEDSRLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEAN 465
R II G+A+G+LYLHEDS IIHRD+KASN+LLD+ KI+DFGMAR+F + +
Sbjct: 684 RFGIITGVAKGLLYLHEDSHNCIIHRDIKASNILLDDKWTAKIADFGMARLFPEDQSQVK 863
Query: 466 TTTIVGTYGYMAPEYAMEGLYSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAW 525
T + GT GYMAPEY M G S+K+DVF +GVL+LE+ITG RN+ F +LL +A+
Sbjct: 864 -TRVAGTNGYMAPEYMMHGRLSVKADVFSYGVLVLELITGQRNSSFNLXVEEHNLLDWAY 1040
Query: 526 HLWNDGKGLELRDPLLLCP--GDQFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMNESVML 583
++ G+ LE+ D L +Q + + LLC+Q D RPTM +V+ L +S
Sbjct: 1041KMYKKGRSLEIVDSALASTVLTEQVDMCIQLALLCIQGDPQLRPTMRRIVVKLSRKS--- 1211
Query: 584 GQPGKP 589
P KP
Sbjct: 1212--PTKP 1223
>TC80060 similar to PIR|D96574|D96574 hypothetical protein T3F20.24
[imported] - Arabidopsis thaliana, partial (32%)
Length = 1547
Score = 264 bits (675), Expect = 7e-71
Identities = 143/277 (51%), Positives = 186/277 (66%), Gaps = 8/277 (2%)
Frame = +1
Query: 308 NFTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVK 367
NF NK+GEGGFGPVYKG+LSDG +A+K+LS S+QG+ EF+NE+ +I LQH NLVK
Sbjct: 190 NFDPKNKIGEGGFGPVYKGVLSDGAVIAVKQLSSKSKQGNREFVNEIGMISALQHPNLVK 369
Query: 368 LLGFCVDGEEKLLVYEYLPNGSLDVVLF---EQHAQLDWTKRLDIINGIARGILYLHEDS 424
L G C++G + LLVYEY+ N SL LF EQ LDW R+ I GIARG+ YLHE+S
Sbjct: 370 LYGCCIEGNQLLLVYEYMENNSLARALFGKPEQRLNLDWRTRMKICVGIARGLAYLHEES 549
Query: 425 RLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANT---TTIVGTYGYMAPEYA 481
RL+I+HRD+KA+NVLLD ++N KISDFG+A++ + E NT T I GT GYMAPEYA
Sbjct: 550 RLKIVHRDIKATNVLLDKNLNAKISDFGLAKL----DEEENTHISTRIAGTIGYMAPEYA 717
Query: 482 MEGLYSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAWHLWNDGKGLELRDPLL 541
M G + K+DV+ FGV+ LEI++G+ N + + LL +A+ L G LEL DP L
Sbjct: 718 MRGYLTDKADVYSFGVVALEIVSGMSNTNYRPKEEFVYLLDWAYVLQEQGNLLELVDPTL 897
Query: 542 --LCPGDQFLRYMNIGLLCVQEDAFDRPTMSSVVLML 576
++ +R + + LLC RP MSSVV ML
Sbjct: 898 GSKYSSEEAMRMLQLALLCTNPSPTLRPPMSSVVSML 1008
>BG453591 similar to PIR|T05181|T05 S-receptor kinase (EC 2.7.1.-) T6K22.120
precursor - Arabidopsis thaliana, partial (24%)
Length = 640
Score = 260 bits (665), Expect = 1e-69
Identities = 127/191 (66%), Positives = 159/191 (82%), Gaps = 3/191 (1%)
Frame = +3
Query: 308 NFTDSNKLGEGGFGPVYKGILS-DGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLV 366
+F++ NKLGEGGFGPVYKG L D +E+A+KRLS S+QG+ EF NEV+L KLQH+NLV
Sbjct: 66 DFSNDNKLGEGGFGPVYKGTLVLDRREIAVKRLSGSSKQGTREFKNEVILCSKLQHRNLV 245
Query: 367 KLLGFCVDGEEKLLVYEYLPNGSLDVVLFEQHAQ--LDWTKRLDIINGIARGILYLHEDS 424
K+LG C+ GEEK+L+YEY+PN SLD LF+Q + LDW+KR +II GIARG++YLH+DS
Sbjct: 246 KVLGCCIQGEEKMLIYEYMPNRSLDSFLFDQAQKKLLDWSKRFNIICGIARGLIYLHQDS 425
Query: 425 RLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEG 484
RL+IIHRDLK SN+LLDNDMNPKISDFG+A+I + E NT +VGT+GYMAPEYA++G
Sbjct: 426 RLRIIHRDLKPSNILLDNDMNPKISDFGLAKICGDDQVEGNTNRVVGTHGYMAPEYAIDG 605
Query: 485 LYSIKSDVFGF 495
L+SIKSDVF F
Sbjct: 606 LFSIKSDVFSF 638
>BG586735 similar to GP|11275527|db putative receptor-like protein kinase
{Oryza sativa (japonica cultivar-group)}, partial (57%)
Length = 794
Score = 180 bits (457), Expect(2) = 2e-67
Identities = 86/145 (59%), Positives = 111/145 (76%)
Frame = +3
Query: 325 KGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVKLLGFCVDGEEKLLVYEY 384
+G++ +G+EVAIK+LS+ S QG EF NEV L+L++QHKNLV LLG C +G EK+LVYEY
Sbjct: 6 RGLMPNGEEVAIKKLSMESRQGIREFTNEVRLLLRIQHKNLVTLLGCCAEGPEKMLVYEY 185
Query: 385 LPNGSLDVVLFEQHAQLDWTKRLDIINGIARGILYLHEDSRLQIIHRDLKASNVLLDNDM 444
LPN SLD LF++ LDW R I+ GIARG+LYLHE++ +IIHRD+KASN+LLD +
Sbjct: 186 LPNKSLDHFLFDKKRSLDWMTRFRIVTGIARGLLYLHEEAPERIIHRDIKASNILLDEKL 365
Query: 445 NPKISDFGMARIFAGSEGEANTTTI 469
NPKISDFG+AR+F G + T I
Sbjct: 366 NPKISDFGLARLFPGEDTHVQTFRI 440
Score = 94.7 bits (234), Expect(2) = 2e-67
Identities = 50/114 (43%), Positives = 71/114 (61%), Gaps = 1/114 (0%)
Frame = +2
Query: 474 GYMAPEYAMEGLYSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAWHLWNDGKG 533
GYMAPEYA+ G S+K+DVF +GVL+LEI++G +N LL+YAW L+ GK
Sbjct: 446 GYMAPEYALRGYLSVKTDVFSYGVLVLEIVSGRKNHDLKLDAEKADLLSYAWKLYQGGKI 625
Query: 534 LELRDPLL-LCPGDQFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMNESVMLGQP 586
++L D + GD+ + +GLLC Q +RP M+SV LML ++S L +P
Sbjct: 626 MDLIDQNIGKYNGDEAAMCIQLGLLCCQASLVERPDMNSVNLMLSSDSFTLPKP 787
>TC88193 similar to PIR|G96602|G96602 probable receptor protein kinase
F14G9.24 [imported] - Arabidopsis thaliana, partial
(20%)
Length = 1815
Score = 244 bits (622), Expect(2) = 1e-66
Identities = 130/298 (43%), Positives = 188/298 (62%), Gaps = 20/298 (6%)
Frame = +3
Query: 264 NNSSKIWMITVIAVGVGLVIIIFICYLCFLRNRQSKLFGDCISV---------------- 307
+ +K+ +I + VGVG V + + + ++ R+ KL+ D +
Sbjct: 384 SKKNKVGLIIGVVVGVGAVCFLAVSSIFYILRRR-KLYNDDDDLVGIDTMPNTFSYYELK 560
Query: 308 ----NFTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHK 363
+F NKLGEGGFGPVYKG L+DG+ VA+K+LSI S QG +FI E+ I +QH+
Sbjct: 561 NATSDFNRDNKLGEGGFGPVYKGTLNDGRFVAVKQLSIGSHQGKSQFIAEIATISAVQHR 740
Query: 364 NLVKLLGFCVDGEEKLLVYEYLPNGSLDVVLFEQHAQLDWTKRLDIINGIARGILYLHED 423
NLVKL G C++G ++LLVYEYL N SLD LF L+W+ R D+ G+ARG+ YLHE+
Sbjct: 741 NLVKLYGCCIEGNKRLLVYEYLENKSLDQALFGNVLFLNWSTRYDVCMGVARGLTYLHEE 920
Query: 424 SRLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAME 483
SRL+I+HRD+KASN+LLD+++ PK+SDFG+A+++ + +T + GT GY+APEYAM
Sbjct: 921 SRLRIVHRDVKASNILLDSELVPKLSDFGLAKLYDDKKTHI-STRVAGTIGYLAPEYAMR 1097
Query: 484 GLYSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAWHLWNDGKGLELRDPLL 541
G + K+DVF FGV+ LE+++G N+ + LL +AW L +L DP L
Sbjct: 1098GRLTEKADVFSFGVVALELVSGRPNSDSSLEEDKMYLLDWAWQLHERNCINDLIDPRL 1271
Score = 28.1 bits (61), Expect(2) = 1e-66
Identities = 14/37 (37%), Positives = 20/37 (53%)
Frame = +1
Query: 555 GLLCVQEDAFDRPTMSSVVLMLMNESVMLGQPGKPPF 591
GLLC Q RP+MS VV ML+ + + +P +
Sbjct: 1315 GLLCTQTSPNLRPSMSRVVAMLLGDIEVSTVTSRPEY 1425
>CA920904 similar to GP|22208478|gb| receptor-like kinase {Sorghum bicolor},
partial (5%)
Length = 560
Score = 246 bits (629), Expect = 1e-65
Identities = 118/118 (100%), Positives = 118/118 (100%)
Frame = -3
Query: 505 GIRNAGFCYSKTTPSLLAYAWHLWNDGKGLELRDPLLLCPGDQFLRYMNIGLLCVQEDAF 564
GIRNAGFCYSKTTPSLLAYAWHLWNDGKGLELRDPLLLCPGDQFLRYMNIGLLCVQEDAF
Sbjct: 558 GIRNAGFCYSKTTPSLLAYAWHLWNDGKGLELRDPLLLCPGDQFLRYMNIGLLCVQEDAF 379
Query: 565 DRPTMSSVVLMLMNESVMLGQPGKPPFSVGRLNFIDQNELDLEEEYSVNFLTVSDILP 622
DRPTMSSVVLMLMNESVMLGQPGKPPFSVGRLNFIDQNELDLEEEYSVNFLTVSDILP
Sbjct: 378 DRPTMSSVVLMLMNESVMLGQPGKPPFSVGRLNFIDQNELDLEEEYSVNFLTVSDILP 205
>BQ136505 similar to PIR|T05754|T05 S-receptor kinase (EC 2.7.1.-) M4I22.110
precursor - Arabidopsis thaliana, partial (23%)
Length = 618
Score = 244 bits (624), Expect = 5e-65
Identities = 126/202 (62%), Positives = 154/202 (75%), Gaps = 2/202 (0%)
Frame = +2
Query: 328 LSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVKLLGFCVDGEEKLLVYEYLPN 387
L G+EVAIKR S S+QG EEF NEV+LI KLQH+NLVKLLG C+ EEKLL+YEY+PN
Sbjct: 11 LIGGKEVAIKRNSKMSDQGLEEFKNEVLLIAKLQHRNLVKLLGCCIHREEKLLIYEYMPN 190
Query: 388 GSLDVVLFEQHAQ--LDWTKRLDIINGIARGILYLHEDSRLQIIHRDLKASNVLLDNDMN 445
SLD +F++ LDW+KR II G+ARG+LYLH+DSRL+IIHRDLK SN+LLD MN
Sbjct: 191 RSLDYFIFDETRSKLLDWSKRSHIIAGVARGLLYLHQDSRLRIIHRDLKLSNILLDALMN 370
Query: 446 PKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEGLYSIKSDVFGFGVLLLEIITG 505
PKISDFG+AR F G + EA T +VGTYGYM PEYA+ G YS+KSDVF FGV++LEII+G
Sbjct: 371 PKISDFGLARTFCGDQVEAKTRKLVGTYGYMPPEYAVHGRYSMKSDVFSFGVIVLEIISG 550
Query: 506 IRNAGFCYSKTTPSLLAYAWHL 527
+ F + +LL +AW L
Sbjct: 551 KKIKVFYDPXHSLNLLGHAWRL 616
>TC92541 similar to PIR|T02153|T02153 protein kinase homolog T1F15.1 -
Arabidopsis thaliana, partial (22%)
Length = 791
Score = 135 bits (339), Expect(3) = 6e-64
Identities = 65/114 (57%), Positives = 88/114 (77%), Gaps = 2/114 (1%)
Frame = +2
Query: 306 SVNFTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNL 365
+++F+ NKLG+GG+GPVYKGIL GQE+A+KRLS S QG EF NE++LI +LQH NL
Sbjct: 11 TIDFSPENKLGQGGYGPVYKGILPTGQEIAVKRLSKTSRQGIVEFKNELVLICELQHTNL 190
Query: 366 VKLLGFCVDGEEKLLVYEYLPNGSLDVVLFEQHAQ--LDWTKRLDIINGIARGI 417
V+LLG C+ EE++L+YEY+ N SLD LF+ + LDW KRL+II GI++G+
Sbjct: 191 VQLLGCCIHEEERILIYEYMSNKSLDFYLFDSTRRKCLDWKKRLNIIEGISQGL 352
Score = 88.6 bits (218), Expect(3) = 6e-64
Identities = 51/111 (45%), Positives = 60/111 (53%), Gaps = 25/111 (22%)
Frame = +1
Query: 451 FGMARIFAGSEGEANTTTIVGT-------------------------YGYMAPEYAMEGL 485
FGMAR+F E NT IVGT GYM+PEYAMEG+
Sbjct: 454 FGMARMFTQQESVVNTNRIVGT**VL*TFLMIT*RKRF*FFK*FFLSSGYMSPEYAMEGI 633
Query: 486 YSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAWHLWNDGKGLEL 536
S KSDV+ FGVLLLEII G RN F +L+ +AW LWNDG+ L+L
Sbjct: 634 CSTKSDVYSFGVLLLEIICGRRNNSFYDVDRPLNLIGHAWELWNDGEYLQL 786
Score = 60.8 bits (146), Expect(3) = 6e-64
Identities = 28/34 (82%), Positives = 32/34 (93%)
Frame = +3
Query: 418 LYLHEDSRLQIIHRDLKASNVLLDNDMNPKISDF 451
LYLH+ SRL+IIHRDLKASN+LLD +MNPKISDF
Sbjct: 354 LYLHKYSRLKIIHRDLKASNILLDENMNPKISDF 455
>TC89703 similar to GP|11994595|dbj|BAB02650. receptor-like serine/threonine
kinase {Arabidopsis thaliana}, partial (26%)
Length = 1212
Score = 172 bits (437), Expect(2) = 5e-63
Identities = 85/143 (59%), Positives = 107/143 (74%), Gaps = 3/143 (2%)
Frame = +2
Query: 308 NFTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVK 367
NF SNK+GEGGFGPVYKG LSDG +A+K LS S+QG+ EF+NE+ +I LQH LVK
Sbjct: 29 NFDISNKIGEGGFGPVYKGRLSDGTLIAVKLLSSKSKQGNREFLNEIGMISALQHPQLVK 208
Query: 368 LLGFCVDGEEKLLVYEYLPNGSLDVVLF---EQHAQLDWTKRLDIINGIARGILYLHEDS 424
L G CV+G++ +L+YEYL N SL LF E +LDW R I GIARG+ YLHE+S
Sbjct: 209 LYGCCVEGDQLMLIYEYLENNSLARALFGPAEHQIRLDWPTRYKICVGIARGLAYLHEES 388
Query: 425 RLQIIHRDLKASNVLLDNDMNPK 447
RL+++HRD+KA+NVLLD D+NPK
Sbjct: 389 RLKVVHRDIKATNVLLDKDLNPK 457
Score = 87.4 bits (215), Expect(2) = 5e-63
Identities = 59/138 (42%), Positives = 76/138 (54%), Gaps = 8/138 (5%)
Frame = +3
Query: 447 KISDFGMARIFAGSEGEANT---TTIVGTYGYMAPEYAMEGLYSIKSDVFGFGVLLLEII 503
KISDFG+A++ + E NT T I GTYGYMAPEYAM G + K+DV+ FG++ LEI+
Sbjct: 456 KISDFGLAKL----DEEENTHISTRIAGTYGYMAPEYAMHGYLTDKADVYSFGIVALEIL 623
Query: 504 TGIRNAGFCYSKTTPSLLAYAWHLWNDGKGLELRDPLLLCPGDQF-----LRYMNIGLLC 558
G N + LL +A L G +EL D L G F + +N+ LLC
Sbjct: 624 HGSNNTILRQKEEAFHLLDWAHILKEKGNEIELVDKRL---GSNFNKEEAMLMINVALLC 794
Query: 559 VQEDAFDRPTMSSVVLML 576
+ RP MSSVV ML
Sbjct: 795 TNVTSSLRPAMSSVVSML 848
>TC80500 similar to PIR|G96602|G96602 probable receptor protein kinase
F14G9.24 [imported] - Arabidopsis thaliana, partial
(13%)
Length = 1015
Score = 230 bits (587), Expect = 1e-60
Identities = 122/256 (47%), Positives = 170/256 (65%), Gaps = 1/256 (0%)
Frame = +1
Query: 325 KGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVKLLGFCVDGEEKLLVYEY 384
+GIL+DG++VA+K+LSI S QG +F+ E+ I +QH+NLVKL G C++G ++LLVYEY
Sbjct: 19 QGILNDGRDVAVKQLSIGSHQGKSQFVAEIATISAVQHRNLVKLYGCCIEGSKRLLVYEY 198
Query: 385 LPNGSLDVVLFEQHAQLDWTKRLDIINGIARGILYLHEDSRLQIIHRDLKASNVLLDNDM 444
L N SLD LF L+W+ R DI G+ARG+ YLHE+SRL+I+HRD+KASN+LLD+++
Sbjct: 199 LENKSLDQALFGNVLFLNWSTRYDICMGVARGLTYLHEESRLRIVHRDVKASNILLDSEL 378
Query: 445 NPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEGLYSIKSDVFGFGVLLLEIIT 504
PKISDFG+A+++ + +T + GT GY+APEYAM G + K+DVF FGV+ LE+++
Sbjct: 379 VPKISDFGLAKLYDDKKTHI-STRVAGTIGYLAPEYAMRGHLTEKADVFSFGVVALELVS 555
Query: 505 GIRNAGFCYSKTTPSLLAYAWHLWNDGKGLELRDPLLL-CPGDQFLRYMNIGLLCVQEDA 563
G N+ LL +AW L EL DP L ++ R + I LLC Q
Sbjct: 556 GRPNSDSTLEGEKMYLLEWAWQLHERNTINELIDPRLSEFNKEEVQRLVGIALLCTQTSP 735
Query: 564 FDRPTMSSVVLMLMNE 579
RP+MS VV ML +
Sbjct: 736 TLRPSMSRVVAMLSGD 783
>TC78270 similar to GP|16649103|gb|AAL24403.1 Unknown protein {Arabidopsis
thaliana}, partial (80%)
Length = 1463
Score = 223 bits (569), Expect = 1e-58
Identities = 122/279 (43%), Positives = 172/279 (60%), Gaps = 5/279 (1%)
Frame = +3
Query: 306 SVNFTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNL 365
S NF+ +NK+GEGGFG VYKG+L G+ AIK LS S+QG +EF+ E+ +I +++H+NL
Sbjct: 300 SDNFSPANKIGEGGFGSVYKGVLKGGKLAAIKVLSTESKQGVKEFLTEINVISEIKHENL 479
Query: 366 VKLLGFCVDGEEKLLVYEYLPNGSLDVVLF---EQHAQLDWTKRLDIINGIARGILYLHE 422
V L G CV+G+ ++LVY YL N SL L + DW R I G+ARG+ +LHE
Sbjct: 480 VILYGCCVEGDHRILVYNYLENNSLSQTLLAGGHSNIYFDWQTRRRICLGVARGLAFLHE 659
Query: 423 DSRLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAM 482
+ I+HRD+KASN+LLD D+ PKISDFG+A++ +T + GT GY+APEYA+
Sbjct: 660 EVLPHIVHRDIKASNILLDKDLTPKISDFGLAKLIPSYMTHV-STRVAGTIGYLAPEYAI 836
Query: 483 EGLYSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAWHLWNDGKGLELRDPLL- 541
G + K+D++ FGVLL+EI++G N +L W L+ + +L D L
Sbjct: 837 RGQLTRKADIYSFGVLLVEIVSGRSNTNTRLPIADQYILETTWQLYERKELAQLVDISLN 1016
Query: 542 -LCPGDQFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMNE 579
++ + + I LLC Q+ RPTMSSVV ML E
Sbjct: 1017GEFDAEEACKILKIALLCTQDTPKLRPTMSSVVKMLTGE 1133
>TC87048 similar to GP|8778594|gb|AAF79602.1| F5M15.3 {Arabidopsis
thaliana}, partial (77%)
Length = 1571
Score = 222 bits (565), Expect = 4e-58
Identities = 118/274 (43%), Positives = 179/274 (65%), Gaps = 6/274 (2%)
Frame = +1
Query: 309 FTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVKL 368
F ++N +GEGGFG V+KG LS G+ VA+K+LS QG +EF+ EV+++ L H NLVKL
Sbjct: 466 FKEANLIGEGGFGKVFKGRLSTGELVAVKQLSHDGRQGFQEFVTEVLMLSLLHHSNLVKL 645
Query: 369 LGFCVDGEEKLLVYEYLPNGSLDVVLFE---QHAQLDWTKRLDIINGIARGILYLHEDSR 425
+G+C DG+++LLVYEY+P GSL+ LF+ L W+ R+ I G ARG+ YLH +
Sbjct: 646 IGYCTDGDQRLLVYEYMPMGSLEDHLFDLPQDKEPLSWSSRMKIAVGAARGLEYLHCKAD 825
Query: 426 LQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEGL 485
+I+RDLK++N+LLD+D +PK+SDFG+A++ + +T ++GTYGY APEYAM G
Sbjct: 826 PPVIYRDLKSANILLDSDFSPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGK 1005
Query: 486 YSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAWHLWND-GKGLELRDPLLL-- 542
++KSD++ FGV+LLE+ITG R +L++++ ++D K + + DPLL
Sbjct: 1006LTLKSDIYSFGVVLLELITGRRAIDASKKPGEQNLVSWSRPYFSDRRKFVHMADPLLQGH 1185
Query: 543 CPGDQFLRYMNIGLLCVQEDAFDRPTMSSVVLML 576
P + + I +C+QE RP + +V+ L
Sbjct: 1186FPVRCLHQAIAITAMCLQEQPKFRPLIGDIVVAL 1287
>TC79033 similar to GP|21554229|gb|AAM63304.1 somatic embryogenesis
receptor-like kinase putative {Arabidopsis thaliana},
partial (77%)
Length = 1762
Score = 215 bits (548), Expect = 4e-56
Identities = 127/303 (41%), Positives = 178/303 (57%), Gaps = 13/303 (4%)
Frame = +1
Query: 308 NFTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVK 367
NF NKLGEGGFG VY G L DG ++A+KRL + S + EF EV ++ +++HKNL+
Sbjct: 460 NFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLKVWSNKADMEFAVEVEILARVRHKNLLS 639
Query: 368 LLGFCVDGEEKLLVYEYLPNGSLDVVLFEQHAQ---LDWTKRLDIINGIARGILYLHEDS 424
L G+C +G+E+L+VY+Y+PN SL L QH+ LDW +R++I G A GI+YLH +
Sbjct: 640 LRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSTESLLDWNRRMNIAIGSAEGIVYLHVQA 819
Query: 425 RLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEG 484
IIHRD+KASNVLLD+D +++DFG A++ TT + GT GY+APEYAM G
Sbjct: 820 TPHIIHRDVKASNVLLDSDFQARVADFGFAKLIPDGATHV-TTRVKGTLGYLAPEYAMLG 996
Query: 485 LYSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAWHLWNDGKGLELRDPLLLCP 544
+ DV+ FG+LLLE+ +G + S ++ +A L + K EL DP L
Sbjct: 997 KANESCDVYSFGILLLELASGKKPLEKLSSSVKRAINDWALPLACEKKFSELADPRL--N 1170
Query: 545 GD----QFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMNES------VMLGQPGKPPFSVG 594
GD + R + + L+C Q RPTM VV +L ES + + K P +VG
Sbjct: 1171GDYVEEELKRVILVALICAQNQPEKRPTMVEVVELLKGESKEKVLQLENNELFKNPLAVG 1350
Query: 595 RLN 597
N
Sbjct: 1351NTN 1359
>BQ155172 weakly similar to GP|6686398|gb F1E22.15 {Arabidopsis thaliana},
partial (10%)
Length = 637
Score = 208 bits (529), Expect(2) = 3e-55
Identities = 105/177 (59%), Positives = 130/177 (73%), Gaps = 2/177 (1%)
Frame = +3
Query: 417 ILYLHEDSRLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYM 476
+LYLH+DSRL++IHRDLK SN+LLD +M PKISDFG+ARI G E EA+T +VGTYGYM
Sbjct: 78 LLYLHQDSRLRVIHRDLKTSNILLDEEMQPKISDFGLARIVGGKETEASTERVVGTYGYM 257
Query: 477 APEYAMEGLYSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAWHLWNDGKGLEL 536
+PEYA++G +S KSDVF FGV+LLEII+G +N GF SK SLL YAW LW + K +L
Sbjct: 258 SPEYALDGYFSTKSDVFSFGVVLLEIISGKKNTGFYRSKEISSLLGYAWRLWREDKLQDL 437
Query: 537 RDPLL--LCPGDQFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMNESVMLGQPGKPPF 591
DP L +QF+R IGLLCVQ++ DRP MS+VV ML NE+ L P +P F
Sbjct: 438 MDPTLSDTYNVNQFIRCSQIGLLCVQDEPDDRPHMSNVVTMLDNETTTLLTPKQPTF 608
Score = 26.2 bits (56), Expect(2) = 3e-55
Identities = 11/16 (68%), Positives = 12/16 (74%)
Frame = +2
Query: 401 LDWTKRLDIINGIARG 416
LDW R +II GIARG
Sbjct: 29 LDWPMRFEIILGIARG 76
>TC93437 similar to GP|11227578|emb|CAC16506. unnamed protein product
{Arabidopsis thaliana}, partial (62%)
Length = 978
Score = 211 bits (537), Expect = 7e-55
Identities = 106/232 (45%), Positives = 155/232 (66%), Gaps = 3/232 (1%)
Frame = +3
Query: 308 NFTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVK 367
+F S K+G GG+G VYKG+L DG +VAIK LS+ S+QG+ EF+ E+ +I +QH NLVK
Sbjct: 246 DFHPSCKIGGGGYGVVYKGVLRDGTQVAIKSLSVESKQGTHEFMTEIAMISNIQHPNLVK 425
Query: 368 LLGFCVDGEEKLLVYEYLPNGSLDVVLFEQHAQ---LDWTKRLDIINGIARGILYLHEDS 424
L+GFC++G ++LVYE+L N SL L ++ LDW KR I G A G+ +LHE++
Sbjct: 426 LIGFCIEGNHRILVYEFLENNSLTSSLLGSKSKCVPLDWQKRAIICRGTASGLSFLHEEA 605
Query: 425 RLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEG 484
+ I+HRD+KASN+LLD + +PKI DFG+A++F + +T + GT GY+APEYA+
Sbjct: 606 QPNIVHRDIKASNILLDENFHPKIGDFGLAKLFPDNVTHV-STRVAGTMGYLAPEYALLR 782
Query: 485 LYSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAWHLWNDGKGLEL 536
+ K+DV+ FG+L+LEII+G + + L+ +AW L + LEL
Sbjct: 783 QLTKKADVYSFGILMLEIISGKSSXKAAFGDNILVLVEWAWKLKEXNRLLEL 938
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.141 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,220,490
Number of Sequences: 36976
Number of extensions: 308257
Number of successful extensions: 3076
Number of sequences better than 10.0: 720
Number of HSP's better than 10.0 without gapping: 2361
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2427
length of query: 622
length of database: 9,014,727
effective HSP length: 102
effective length of query: 520
effective length of database: 5,243,175
effective search space: 2726451000
effective search space used: 2726451000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC123975.9


