

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123975.11 - phase: 0
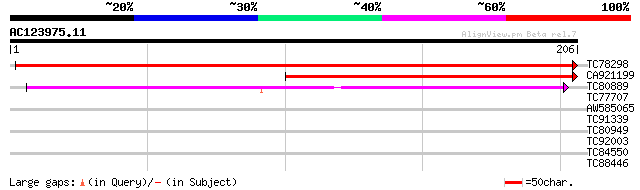
(206 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC78298 similar to PIR|T51950|T51950 DNA-directed RNA polymerase... 395 e-111
CA921199 similar to PIR|T51950|T51 DNA-directed RNA polymerase (... 211 2e-55
TC80889 weakly similar to GP|8896071|gb|AAF81222.1| RPB5d {Brass... 142 1e-34
TC77707 weakly similar to GP|8777405|dbj|BAA96995.1 SCARECROW ge... 30 0.85
AW585065 similar to GP|17130377|dbj general secretion pathway pr... 28 3.2
TC91339 homologue to PIR|B84609|B84609 hypothetical protein At2g... 27 4.2
TC80949 similar to PIR|T52422|T52422 alternative oxidase-related... 27 5.5
TC92003 weakly similar to GP|17978838|gb|AAL47552.1 IAA-amino ac... 27 5.5
TC84550 weakly similar to PIR|T46095|T46095 hypothetical protein... 27 7.2
TC88446 weakly similar to GP|15810469|gb|AAL07122.1 unknown prot... 26 9.4
>TC78298 similar to PIR|T51950|T51950 DNA-directed RNA polymerase (EC
2.7.7.6) 23K chain [imported] - Arabidopsis thaliana,
partial (96%)
Length = 860
Score = 395 bits (1015), Expect = e-111
Identities = 198/204 (97%), Positives = 200/204 (97%)
Frame = +3
Query: 3 FSEEEITRLYRIRKTVMQMLKDRNYLVGDFELNMSKHDFKDKYGENMKREDLVINKTKKD 62
F ++ RLYRIRKTVMQMLKDRNYLVGDFELNMSKHDFKDKYGENMKREDLVINKTKKD
Sbjct: 45 FQRKK*PRLYRIRKTVMQMLKDRNYLVGDFELNMSKHDFKDKYGENMKREDLVINKTKKD 224
Query: 63 KPSDQIYVFFPEEAKVGVKTMKTYTNRMNSENVYRAILVCQTSLTPFAKTCVSEIASKFH 122
KPSDQIYVFFPEEAKVGVKTMKTYTNRMNSENVYRAILVCQTSLTPFAKTCVSEIASKFH
Sbjct: 225 KPSDQIYVFFPEEAKVGVKTMKTYTNRMNSENVYRAILVCQTSLTPFAKTCVSEIASKFH 404
Query: 123 LEVFQEAELLVNIKEHVLVPEHQILNDTEKKTLLERYTVKETQLPRIQVTDPVARYYGLK 182
LEVFQEAELLVNIKEHVLVPEHQILNDTEKKTLLERYTVKETQLPRIQVTDPVARYYGLK
Sbjct: 405 LEVFQEAELLVNIKEHVLVPEHQILNDTEKKTLLERYTVKETQLPRIQVTDPVARYYGLK 584
Query: 183 RGQVVKIIRPSETAGRYVTYRFVV 206
RGQVVKIIRPSETAGRYVTYRFVV
Sbjct: 585 RGQVVKIIRPSETAGRYVTYRFVV 656
>CA921199 similar to PIR|T51950|T51 DNA-directed RNA polymerase (EC 2.7.7.6)
23K chain [imported] - Arabidopsis thaliana, partial
(49%)
Length = 756
Score = 211 bits (536), Expect = 2e-55
Identities = 105/106 (99%), Positives = 106/106 (99%)
Frame = -1
Query: 101 VCQTSLTPFAKTCVSEIASKFHLEVFQEAELLVNIKEHVLVPEHQILNDTEKKTLLERYT 160
+CQTSLTPFAKTCVSEIASKFHLEVFQEAELLVNIKEHVLVPEHQILNDTEKKTLLERYT
Sbjct: 483 LCQTSLTPFAKTCVSEIASKFHLEVFQEAELLVNIKEHVLVPEHQILNDTEKKTLLERYT 304
Query: 161 VKETQLPRIQVTDPVARYYGLKRGQVVKIIRPSETAGRYVTYRFVV 206
VKETQLPRIQVTDPVARYYGLKRGQVVKIIRPSETAGRYVTYRFVV
Sbjct: 303 VKETQLPRIQVTDPVARYYGLKRGQVVKIIRPSETAGRYVTYRFVV 166
>TC80889 weakly similar to GP|8896071|gb|AAF81222.1| RPB5d {Brassica napus},
partial (83%)
Length = 953
Score = 142 bits (357), Expect = 1e-34
Identities = 79/198 (39%), Positives = 119/198 (59%), Gaps = 1/198 (0%)
Frame = +3
Query: 7 EITRLYRIRKTVMQMLKDRNYLVGDFELNMSKHDFKDKYGENMKREDLVINKTKKDKPSD 66
E R Y R+TV++MLKDR Y + E+ +S DF+ +G++ + L + T PS
Sbjct: 147 ESHRYYLSRRTVLEMLKDRGYSIPSDEIQLSLDDFRQIHGQSPDVDRLRLTATHATNPSK 326
Query: 67 QIYVFFPEEAKVGVKTMKTYTNRM-NSENVYRAILVCQTSLTPFAKTCVSEIASKFHLEV 125
+I V F V V ++ ++ N E++ IL+ Q +T A V+ ++ F +E+
Sbjct: 327 RILVVFSGPGIVKVNGVRDIAGQIVNRESLTGLILIVQNQITSQALKAVNLLS--FKVEI 500
Query: 126 FQEAELLVNIKEHVLVPEHQILNDTEKKTLLERYTVKETQLPRIQVTDPVARYYGLKRGQ 185
FQ +LLVN +HVL P+HQ+L D +KK LL++Y ++E QLPR+ TD +ARYYGL+RGQ
Sbjct: 501 FQITDLLVNATKHVLKPKHQVLTDKQKKNLLKKYDIQEKQLPRMLQTDAIARYYGLQRGQ 680
Query: 186 VVKIIRPSETAGRYVTYR 203
VVK+ E +VTYR
Sbjct: 681 VVKVTYTGEITQMHVTYR 734
>TC77707 weakly similar to GP|8777405|dbj|BAA96995.1 SCARECROW gene
regulator-like {Arabidopsis thaliana}, partial (65%)
Length = 2161
Score = 29.6 bits (65), Expect = 0.85
Identities = 15/58 (25%), Positives = 33/58 (56%), Gaps = 3/58 (5%)
Frame = +2
Query: 136 KEHVLVPEHQILNDTEKKTLLERYTVKETQLPRIQVT---DPVARYYGLKRGQVVKII 190
++H+ + + QI T+ TLL+ + P +++T DPV++Y RG+ ++++
Sbjct: 1181 EDHIHIIDFQIAQGTQWMTLLQALAARPGGAPHVRITGIDDPVSKY---ARGKGLEVV 1345
>AW585065 similar to GP|17130377|dbj general secretion pathway protein D
{Nostoc sp. PCC 7120}, partial (2%)
Length = 413
Score = 27.7 bits (60), Expect = 3.2
Identities = 12/25 (48%), Positives = 15/25 (60%)
Frame = +2
Query: 133 VNIKEHVLVPEHQILNDTEKKTLLE 157
+ IK H L P+H LND K+ LE
Sbjct: 323 IGIKSHTLNPDHGALNDDTKEPPLE 397
>TC91339 homologue to PIR|B84609|B84609 hypothetical protein At2g22120
[imported] - Arabidopsis thaliana, partial (47%)
Length = 681
Score = 27.3 bits (59), Expect = 4.2
Identities = 9/30 (30%), Positives = 19/30 (63%)
Frame = -3
Query: 39 HDFKDKYGENMKREDLVINKTKKDKPSDQI 68
H F+DK G R+D + +++KD+ +++
Sbjct: 259 HQFRDKSGTEFVRKDRLRGRSRKDRREEEV 170
>TC80949 similar to PIR|T52422|T52422 alternative oxidase-related protein
IMMUTANS [validated] - Arabidopsis thaliana, partial
(69%)
Length = 1369
Score = 26.9 bits (58), Expect = 5.5
Identities = 10/28 (35%), Positives = 19/28 (67%)
Frame = -2
Query: 92 SENVYRAILVCQTSLTPFAKTCVSEIAS 119
+E+V + + C T +TPF++ V E++S
Sbjct: 366 TESVRKILTDCSTLITPFSQALVDEVSS 283
>TC92003 weakly similar to GP|17978838|gb|AAL47552.1 IAA-amino acid
conjugate hydrolase-like protein {Arabidopsis thaliana},
partial (35%)
Length = 610
Score = 26.9 bits (58), Expect = 5.5
Identities = 20/67 (29%), Positives = 29/67 (42%), Gaps = 3/67 (4%)
Frame = -3
Query: 81 KTMKTYTNRMNSENVYRAILVCQT---SLTPFAKTCVSEIASKFHLEVFQEAELLVNIKE 137
K K+ TN N+ + L+ T LTP+ +TC+ KF +L+N
Sbjct: 323 KRSKSRTNFSNNTSFSHRPLINNT*LVKLTPYNRTCLEFFICKF--------RVLMNFSS 168
Query: 138 HVLVPEH 144
VL P H
Sbjct: 167 DVLHPFH 147
>TC84550 weakly similar to PIR|T46095|T46095 hypothetical protein T25B15.20
- Arabidopsis thaliana, partial (1%)
Length = 969
Score = 26.6 bits (57), Expect = 7.2
Identities = 16/69 (23%), Positives = 28/69 (40%)
Frame = +1
Query: 57 NKTKKDKPSDQIYVFFPEEAKVGVKTMKTYTNRMNSENVYRAILVCQTSLTPFAKTCVSE 116
NKT +D PSD + F E + ++ ++N VC ++ + C
Sbjct: 598 NKTDEDLPSDALNTFHDESNPLEATSLSA---KLNESREISGTEVCLENVDVASVACAIN 768
Query: 117 IASKFHLEV 125
+ SK +V
Sbjct: 769 VESKLGSDV 795
>TC88446 weakly similar to GP|15810469|gb|AAL07122.1 unknown protein
{Arabidopsis thaliana}, partial (64%)
Length = 1435
Score = 26.2 bits (56), Expect = 9.4
Identities = 16/44 (36%), Positives = 24/44 (54%), Gaps = 2/44 (4%)
Frame = -3
Query: 27 YLVGDFELNMSKHDFKDKYGENMKR--EDLVINKTKKDKPSDQI 68
++VG+F LN+ K + K+ D++ K KKD PSD I
Sbjct: 1355 FIVGNFFLNLRKEANSKSIPKLYKKLTVDIISVKLKKDIPSDII 1224
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.135 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,864,501
Number of Sequences: 36976
Number of extensions: 56774
Number of successful extensions: 304
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 303
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 303
length of query: 206
length of database: 9,014,727
effective HSP length: 92
effective length of query: 114
effective length of database: 5,612,935
effective search space: 639874590
effective search space used: 639874590
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC123975.11


