

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123898.9 + phase: 0 /pseudo
(194 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
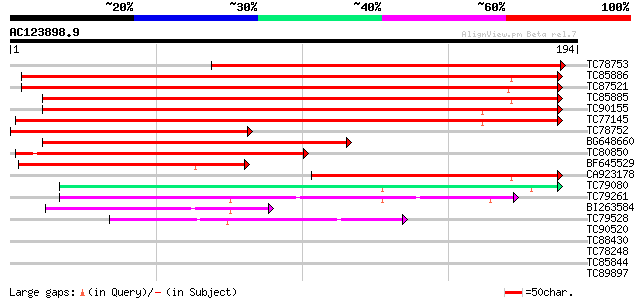
Score E
Sequences producing significant alignments: (bits) Value
TC78753 similar to PIR|E96751|E96751 hypothetical protein F28P22... 221 1e-58
TC85886 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 189 4e-49
TC87521 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 187 3e-48
TC85885 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 182 5e-47
TC90155 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 175 9e-45
TC77145 homologue to SP|P31656|CADH_MEDSA Cinnamyl-alcohol dehyd... 167 3e-42
TC78752 similar to GP|22857590|gb|AAN09864.1 putative cinnamyl-a... 166 7e-42
BG648660 similar to SP|Q9ZRF1|MTD_ Probable mannitol dehydrogena... 128 1e-30
TC80850 homologue to SP|O82515|MTD_MEDSA Probable mannitol dehyd... 124 3e-29
BF645529 weakly similar to GP|22475166|gb putative sinapyl alcoh... 91 4e-19
CA923178 weakly similar to SP|P93257|MTD_ Probable mannitol dehy... 75 1e-14
TC79080 similar to GP|10177043|dbj|BAB10455. alcohol dehydrogena... 55 2e-08
TC79261 similar to PIR|T49047|T49047 quinone reductase-like prot... 52 1e-07
BI263584 similar to SP|Q9P6C8|ADH1 Alcohol dehydrogenase I (EC 1... 48 2e-06
TC79528 similar to GP|15808674|gb|AAL06644.1 putative quinone ox... 41 3e-04
TC90520 weakly similar to PIR|T08581|T08581 cinnamyl-alcohol deh... 35 0.018
TC88430 weakly similar to PIR|T10824|T10824 auxin-induced protei... 35 0.024
TC78248 similar to GP|16323192|gb|AAL15330.1 At2g47330/T8I13.17 ... 34 0.041
TC85844 homologue to SP|P12886|ADH1_PEA Alcohol dehydrogenase 1 ... 33 0.070
TC89897 similar to GP|17978983|gb|AAL47452.1 At1g79870/F19K16_17... 33 0.091
>TC78753 similar to PIR|E96751|E96751 hypothetical protein F28P22.13
[imported] - Arabidopsis thaliana, partial (47%)
Length = 865
Score = 221 bits (563), Expect = 1e-58
Identities = 114/121 (94%), Positives = 116/121 (95%)
Frame = +3
Query: 70 GVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEEMRALAKT 129
GVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALS LGADQFVVSSNQE+MRALAK+
Sbjct: 3 GVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSLLGADQFVVSSNQEDMRALAKS 182
Query: 130 LDFIIDTASGDHPFDPYMSLLKISGVLALVGFPSEVKFSPASLNLGSRTVAGSAAGGTKE 189
LDFIIDTASGDH FDPYMSLLKISGVL LVGFPSEVKFSPASLNLGSRTVAGS GGTKE
Sbjct: 183 LDFIIDTASGDHLFDPYMSLLKISGVLVLVGFPSEVKFSPASLNLGSRTVAGSVTGGTKE 362
Query: 190 I 190
I
Sbjct: 363 I 365
>TC85886 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (94%)
Length = 1395
Score = 189 bits (481), Expect = 4e-49
Identities = 100/186 (53%), Positives = 128/186 (68%), Gaps = 1/186 (0%)
Frame = +2
Query: 5 TFNCVDYDGTITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNMNQ 64
TFN DGTIT GG+S S+V +E + IP S PL AGPLLCAG+TVYSP+ +++
Sbjct: 437 TFNGKSRDGTITYGGFSDSMVADEHFVIRIPDSLPLDGAGPLLCAGVTVYSPLRHFGLDK 616
Query: 65 PGKSLGVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEEMR 124
PG ++GVVGLGGLGHMAVKF KAFG +VTV STS K++EA+ LGAD F+VS + E+M+
Sbjct: 617 PGMNIGVVGLGGLGHMAVKFAKAFGAKVTVISTSPKKEKEAIEHLGADSFLVSRDPEQMQ 796
Query: 125 ALAKTLDFIIDTASGDHPFDPYMSLLKISGVLALVGFPSEVKFSPA-SLNLGSRTVAGSA 183
A TL+ IIDT S HP P + LLK +G L +VG ++ P SL G +++AGS
Sbjct: 797 AATSTLNGIIDTVSASHPVVPLIGLLKSNGKLVMVGAVAKPLELPIFSLLGGRKSIAGSL 976
Query: 184 AGGTKE 189
GG KE
Sbjct: 977 IGGIKE 994
>TC87521 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (98%)
Length = 1433
Score = 187 bits (474), Expect = 3e-48
Identities = 96/186 (51%), Positives = 127/186 (67%), Gaps = 1/186 (0%)
Frame = +2
Query: 5 TFNCVDYDGTITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNMNQ 64
T++ DG+IT GGYS S+V +E + IP PL SA PLLCAGITVYSP+ +++
Sbjct: 434 TYSAKYSDGSITYGGYSDSMVADEHFIVHIPDGLPLESAAPLLCAGITVYSPLRYFGLDK 613
Query: 65 PGKSLGVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEEMR 124
PG ++G+VGLGGLGH+ VKF KAFG VTV STS +K++EA+ LGAD F++S +Q++M+
Sbjct: 614 PGMNIGIVGLGGLGHLGVKFAKAFGANVTVISTSPNKEKEAIENLGADSFLISHDQDKMQ 793
Query: 125 ALAKTLDFIIDTASGDHPFDPYMSLLKISGVLALVGFPSEVKFSP-ASLNLGSRTVAGSA 183
A TLD IIDT S DHP P + LLK G L +VG P + P L +G +T++GS
Sbjct: 794 AAMGTLDGIIDTVSADHPLLPLVGLLKYHGKLVMVGAPDKPPELPHIPLIMGRKTISGSG 973
Query: 184 AGGTKE 189
GG KE
Sbjct: 974 IGGMKE 991
>TC85885 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (97%)
Length = 1330
Score = 182 bits (463), Expect = 5e-47
Identities = 97/179 (54%), Positives = 124/179 (69%), Gaps = 1/179 (0%)
Frame = +3
Query: 12 DGTITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNMNQPGKSLGV 71
DG++T GGYS S+V +E + IP S PL AGPLLCAG+TVYSP+ +++PG ++GV
Sbjct: 456 DGSVTYGGYSDSMVADEHFVIRIPDSLPLDVAGPLLCAGVTVYSPLRHFQLDKPGMNIGV 635
Query: 72 VGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEEMRALAKTLD 131
VGLGGLGHMAVKF KAFG VTV STS SK++EA+ LGAD F+VS + ++M+A TL+
Sbjct: 636 VGLGGLGHMAVKFAKAFGANVTVISTSPSKEKEAIEHLGADSFLVSRDPDQMQAAMGTLN 815
Query: 132 FIIDTASGDHPFDPYMSLLKISGVLALVGFPSEVKFSPA-SLNLGSRTVAGSAAGGTKE 189
IIDT S HP P + LLK +G L +VG ++ P SL G + VAGS GG KE
Sbjct: 816 GIIDTVSASHPILPLIGLLKSNGKLVMVGGVAKPLELPVFSLLGGRKLVAGSLIGGIKE 992
>TC90155 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (78%)
Length = 1051
Score = 175 bits (444), Expect = 9e-45
Identities = 91/179 (50%), Positives = 121/179 (66%), Gaps = 1/179 (0%)
Frame = +2
Query: 12 DGTITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNMNQPGKSLGV 71
DGT+T GGYS S+V +E + IP + PL AGPLLCAG+TVYSP+ +++PG +GV
Sbjct: 167 DGTVTYGGYSDSMVADEHFVIRIPDNIPLEFAGPLLCAGVTVYSPLRFFGLDKPGLHIGV 346
Query: 72 VGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEEMRALAKTLD 131
VGLGGLGHMAVKF KAFG VTV STS +K++EA+ LGAD F++SS+ ++++ TLD
Sbjct: 347 VGLGGLGHMAVKFAKAFGANVTVISTSPNKEKEAIEHLGADSFLISSDPKQIQGAIGTLD 526
Query: 132 FIIDTASGDHPFDPYMSLLKISGVLALVG-FPSEVKFSPASLNLGSRTVAGSAAGGTKE 189
IIDT S HP P + LLK G L ++G ++ +L G + +AGS GG KE
Sbjct: 527 GIIDTVSAVHPLLPMIGLLKSHGKLIMLGVIVQPLQLPEYTLIQGRKILAGSQVGGLKE 703
>TC77145 homologue to SP|P31656|CADH_MEDSA Cinnamyl-alcohol dehydrogenase
(EC 1.1.1.195) (CAD). [Alfalfa] {Medicago sativa},
complete
Length = 1384
Score = 167 bits (422), Expect = 3e-42
Identities = 86/188 (45%), Positives = 121/188 (63%), Gaps = 1/188 (0%)
Frame = +1
Query: 3 VYTFNCVDYDGTITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNM 62
++++N V DG IT+GG++ S VV +++ IP+ PLLCAG+TVYSP+ +
Sbjct: 406 IWSYNDVYTDGKITQGGFAESTVVEQKFVVKIPEGLAPEQVAPLLCAGVTVYSPLSHFGL 585
Query: 63 NQPGKSLGVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEE 122
PG G++GLGG+GHM VK KAFG VTV S+S KK+EAL LGAD ++VSS+
Sbjct: 586 KTPGLRGGILGLGGVGHMGVKVAKAFGHHVTVISSSDKKKKEALEDLGADSYLVSSDTVG 765
Query: 123 MRALAKTLDFIIDTASGDHPFDPYMSLLKISGVLALVG-FPSEVKFSPASLNLGSRTVAG 181
M+ A +LD+IIDT HP +PY+SLLKI G L L+G + ++F + LG +++ G
Sbjct: 766 MQEAADSLDYIIDTVPVGHPLEPYLSLLKIDGKLILMGVINTPLQFVTPMVMLGRKSITG 945
Query: 182 SAAGGTKE 189
S G KE
Sbjct: 946 SFVGSVKE 969
>TC78752 similar to GP|22857590|gb|AAN09864.1 putative cinnamyl-alcohol
dehydrogenase {Oryza sativa (japonica cultivar-group)},
partial (50%)
Length = 668
Score = 166 bits (419), Expect = 7e-42
Identities = 79/83 (95%), Positives = 82/83 (98%)
Frame = +2
Query: 1 GSVYTFNCVDYDGTITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRH 60
GSVYTFN VDYDGTITKGGYSTSIVV+ERYCFLIPKS+PLASAGPLLCAGITVYSPM+RH
Sbjct: 392 GSVYTFNGVDYDGTITKGGYSTSIVVHERYCFLIPKSHPLASAGPLLCAGITVYSPMIRH 571
Query: 61 NMNQPGKSLGVVGLGGLGHMAVK 83
NMNQPGKSLGVVGLGGLGHMAVK
Sbjct: 572 NMNQPGKSLGVVGLGGLGHMAVK 640
>BG648660 similar to SP|Q9ZRF1|MTD_ Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (64%)
Length = 721
Score = 128 bits (322), Expect = 1e-30
Identities = 63/106 (59%), Positives = 77/106 (72%)
Frame = +1
Query: 12 DGTITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNMNQPGKSLGV 71
DGT T GG+S S+V +E + F IP PL +A PLLCAGITVYSP+ +++P ++GV
Sbjct: 403 DGTSTHGGFSDSMVSDEHFVFRIPDQLPLDAAAPLLCAGITVYSPLRHFGLDKPDMNIGV 582
Query: 72 VGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVS 117
VGLGGLGHMAVKF KAFG VTV STS K+ EAL L AD F++S
Sbjct: 583 VGLGGLGHMAVKFAKAFGANVTVISTSPKKENEALEHLRADSFLIS 720
>TC80850 homologue to SP|O82515|MTD_MEDSA Probable mannitol dehydrogenase
(EC 1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Alfalfa], partial (61%)
Length = 710
Score = 124 bits (310), Expect = 3e-29
Identities = 60/100 (60%), Positives = 75/100 (75%)
Frame = +1
Query: 3 VYTFNCVDYDGTITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNM 62
V+T+N Y GT T GGYS +VV++RY P + PL + PLLCAGITVYSPM + M
Sbjct: 412 VFTYNS-PYKGTRTHGGYSDFVVVHQRYVVQFPDNLPLDAGAPLLCAGITVYSPMKYYGM 588
Query: 63 NQPGKSLGVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKK 102
+PGK LGV GLGGLGH+A+KFGKAFGL+VTV +TS +K+
Sbjct: 589 TEPGKHLGVAGLGGLGHVAIKFGKAFGLKVTVITTSPNKE 708
>BF645529 weakly similar to GP|22475166|gb putative sinapyl alcohol
dehydrogenase {Populus tremula x Populus tremuloides},
partial (40%)
Length = 658
Score = 90.5 bits (223), Expect = 4e-19
Identities = 44/80 (55%), Positives = 59/80 (73%), Gaps = 1/80 (1%)
Frame = +3
Query: 4 YTFNCVDYDGTITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNM- 62
+ +N + +DG+IT GGYS +VV+ RY IP+S P+ +A PLLCAGITV+SP+ H +
Sbjct: 390 FVYNGIFWDGSITYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHXLV 569
Query: 63 NQPGKSLGVVGLGGLGHMAV 82
+ GK +GVVGLG LGHMAV
Sbjct: 570 STAGKRIGVVGLGXLGHMAV 629
>CA923178 weakly similar to SP|P93257|MTD_ Probable mannitol dehydrogenase
(EC 1.1.1.255) (NAD-dependent mannitol dehydrogenase).,
partial (34%)
Length = 668
Score = 75.5 bits (184), Expect = 1e-14
Identities = 38/87 (43%), Positives = 55/87 (62%), Gaps = 1/87 (1%)
Frame = -3
Query: 104 EALSFLGADQFVVSSNQEEMRALAKTLDFIIDTASGDHPFDPYMSLLKISGVLALVGFPS 163
EA LGAD F++S+N ++++A ++LDFI+DT S DH P + LLK++G L +VG P
Sbjct: 663 EAKERLGADDFIISTNPDQLQAAKRSLDFILDTVSADHALLPILELLKVNGTLFIVGAPD 484
Query: 164 EVKFSPA-SLNLGSRTVAGSAAGGTKE 189
+ PA L G R++ G GG KE
Sbjct: 483 KPLQLPAFPLIFGKRSIKGGIIGGIKE 403
>TC79080 similar to GP|10177043|dbj|BAB10455. alcohol dehydrogenase-like
protein {Arabidopsis thaliana}, partial (96%)
Length = 1524
Score = 54.7 bits (130), Expect = 2e-08
Identities = 41/180 (22%), Positives = 70/180 (38%), Gaps = 8/180 (4%)
Frame = +3
Query: 18 GGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNMNQPGKSLGVVGLGGL 77
GG + VV ++P S P + L CA T Y M +PG ++ V+G GG+
Sbjct: 657 GGLAEYCVVPANALAVLPNSMPYTESAILGCAVFTAYGAMAHAAEVRPGDTVAVIGTGGV 836
Query: 78 GHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEEMRAL-----AKTLDF 132
G ++ +AFG + +K E LGA + S+ ++ + + K +D
Sbjct: 837 GSSCLQIARAFGASDIIAVDVQDEKLEKAKTLGATHTINSAKEDPIEKILEITGGKGVDV 1016
Query: 133 IIDTASGDHPFDPYMSLLKISGVLALVGFPSEVKFSPASLNLGSR---TVAGSAAGGTKE 189
++ F +K G ++G +N R V GS G ++
Sbjct: 1017AVEALGRPQTFAQCTQSVKDGGKAVMIGLAQAGSLGEVDINRLVRRKIKVIGSYGGRARQ 1196
>TC79261 similar to PIR|T49047|T49047 quinone reductase-like protein -
Arabidopsis thaliana, partial (95%)
Length = 1239
Score = 52.0 bits (123), Expect = 1e-07
Identities = 46/167 (27%), Positives = 78/167 (46%), Gaps = 10/167 (5%)
Frame = +3
Query: 18 GGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNMNQPGKSLGVVGL-GG 76
G Y+ +VV++ F +P+ L +AG L A T + ++ + G+ L V+G GG
Sbjct: 330 GSYAQYLVVDQTQLFRVPEGCDLVAAGALAVAFGTSHVGLVHRAQLKSGQVLLVLGAAGG 509
Query: 77 LGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEEMRAL--------AK 128
+G AV+ GKA G V + ++K + L +G D V N+ ++ K
Sbjct: 510 VGLAAVQIGKACGAIVIAVARG-AEKVQLLKSMGVDHVVDLGNENVTESVKEFLKVKRLK 686
Query: 129 TLDFIIDTASGDHPFDPYMSLLKISGVLALVGFPS-EVKFSPASLNL 174
+D + D G + LLK + ++GF S E+ PA++ L
Sbjct: 687 GIDVLYDPVGG-KLMKESLRLLKWGANILIIGFASGEIPVIPANIAL 824
>BI263584 similar to SP|Q9P6C8|ADH1 Alcohol dehydrogenase I (EC 1.1.1.1).
{Neurospora crassa}, partial (57%)
Length = 682
Score = 48.1 bits (113), Expect = 2e-06
Identities = 27/79 (34%), Positives = 42/79 (52%), Gaps = 1/79 (1%)
Frame = +2
Query: 13 GTITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNMNQPGKSLGVV 72
G G + + + IP+ L S P+LCAGITVY + + + G+S+ +V
Sbjct: 425 GYTVDGSFQQYAIAKAIHVARIPEECDLESISPILCAGITVYKGLKESGV-KAGQSIAIV 601
Query: 73 GL-GGLGHMAVKFGKAFGL 90
G GGLG +A ++ KA G+
Sbjct: 602 GAGGGLGSIAXQYCKAMGI 658
>TC79528 similar to GP|15808674|gb|AAL06644.1 putative quinone
oxidoreductase {Fragaria x ananassa}, partial (72%)
Length = 834
Score = 40.8 bits (94), Expect = 3e-04
Identities = 34/103 (33%), Positives = 48/103 (46%), Gaps = 1/103 (0%)
Frame = +2
Query: 35 PKSYPLASAGPLLCAGITVYSPMMRHNMNQPGKSLGVVG-LGGLGHMAVKFGKAFGLRVT 93
PK+ A A L T Y + R + GKS+ V+G GG+G ++ K
Sbjct: 449 PKNLSFAEAASLPLTIETAYEGLERTGFSA-GKSILVLGGAGGVGTHVIQLAKHVYGASK 625
Query: 94 VFSTSMSKKEEALSFLGADQFVVSSNQEEMRALAKTLDFIIDT 136
V +TS +KK E LS LGAD + +E L + D + DT
Sbjct: 626 VAATSSTKKLELLSNLGAD-LPIDYTKENFEDLPEKFDVVFDT 751
>TC90520 weakly similar to PIR|T08581|T08581 cinnamyl-alcohol dehydrogenase
(EC 1.1.1.195) CAD1 - Arabidopsis thaliana, partial
(41%)
Length = 618
Score = 35.0 bits (79), Expect = 0.018
Identities = 19/44 (43%), Positives = 26/44 (58%), Gaps = 1/44 (2%)
Frame = +1
Query: 14 TITKGGYSTSIVVNERYCFL-IPKSYPLASAGPLLCAGITVYSP 56
T+ G S +V C+ I +S P+ +A PLLCAGITV+ P
Sbjct: 406 TMVFSGMVASPMVATHKCWS*ITESLPMDAAAPLLCAGITVFXP 537
Score = 33.9 bits (76), Expect = 0.041
Identities = 27/77 (35%), Positives = 39/77 (50%), Gaps = 1/77 (1%)
Frame = +3
Query: 6 FNCVDYDGTITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSP-MMRHNMNQ 64
+N + +DG+IT GGYS +VV+ R K S+ LC ++ S ++
Sbjct: 405 YNGIFWDGSITYGGYSQMLVVDYR------KPTNGCSSTSTLCWNNSIXSL*RTTVLVST 566
Query: 65 PGKSLGVVGLGGLGHMA 81
GK L + LGGLG MA
Sbjct: 567 AGKKLVXLVLGGLGXMA 617
>TC88430 weakly similar to PIR|T10824|T10824 auxin-induced protein (clone
MII-3) - mung bean, partial (41%)
Length = 1450
Score = 34.7 bits (78), Expect = 0.024
Identities = 37/137 (27%), Positives = 55/137 (40%), Gaps = 1/137 (0%)
Frame = +2
Query: 1 GSVYTFNCVDYDGTITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRH 60
G++ FN ++ + G + IVV E PK A L A T
Sbjct: 287 GNIQDFNSMEKPKQL--GTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQTAIEGFKTG 460
Query: 61 NMNQPGKSLGVVGLGGLGHMAVKFGK-AFGLRVTVFSTSMSKKEEALSFLGADQFVVSSN 119
+ + V G GG+G + ++ K FG V S S K + F GAD+ VV
Sbjct: 461 DFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSCSTPKLKFVKQF-GADK-VVDYT 634
Query: 120 QEEMRALAKTLDFIIDT 136
+ + + + DFI DT
Sbjct: 635 KTKYEDIEEKFDFIYDT 685
>TC78248 similar to GP|16323192|gb|AAL15330.1 At2g47330/T8I13.17 {Arabidopsis
thaliana}, partial (79%)
Length = 2745
Score = 33.9 bits (76), Expect = 0.041
Identities = 32/105 (30%), Positives = 47/105 (44%), Gaps = 16/105 (15%)
Frame = +2
Query: 56 PMMRHNMNQPG--KSLGVVGL-----GGLGHM----AVKFGKAFGLRVTVFSTSMSKKEE 104
PM+ H M+QP K G +G+ L H A KF KA+G+RV+ MSK E+
Sbjct: 1067 PMIVHIMDQPELQKEEGPIGVICAPTRELAHQIYLEAKKFAKAYGIRVSAVYGGMSKLEQ 1246
Query: 105 ALSFLGADQFVVSS-----NQEEMRALAKTLDFIIDTASGDHPFD 144
+ VV++ + +M+ALA + D FD
Sbjct: 1247 FKELKAGCEIVVATPGRLIDMLKMKALAMLRATYLVLDEADRMFD 1381
>TC85844 homologue to SP|P12886|ADH1_PEA Alcohol dehydrogenase 1 (EC
1.1.1.1). [Garden pea] {Pisum sativum}, complete
Length = 1502
Score = 33.1 bits (74), Expect = 0.070
Identities = 42/179 (23%), Positives = 71/179 (39%), Gaps = 10/179 (5%)
Frame = +2
Query: 20 YSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNMNQPGKSLGVVGLGGLGH 79
+S VV+ I PL L C T + +PG S+ + GLG +G
Sbjct: 536 FSEYTVVHAGCVAKINPDAPLDKVCILSCGICTGLGATINVAKPKPGSSVAIFGLGAVGL 715
Query: 80 MAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEE--MRALAKTLDFIID-- 135
A + + G + +S + E G ++FV + ++ + +A+ + +D
Sbjct: 716 AAAEGARISGASRIIGVDLVSSRFELAKKFGVNEFVNPKDHDKPVQQVIAEMTNGGVDRA 895
Query: 136 ---TASGDHPFDPYMSLLKISGVLALVGFPSE---VKFSPASLNLGSRTVAGSAAGGTK 188
T S + + GV LVG P++ K P +L L RT+ G+ G K
Sbjct: 896 VECTGSIQAMISAFECVHDGWGVAVLVGVPNKDDAFKTHPMNL-LNERTLKGTFYGNYK 1069
>TC89897 similar to GP|17978983|gb|AAL47452.1 At1g79870/F19K16_17
{Arabidopsis thaliana}, partial (53%)
Length = 1100
Score = 32.7 bits (73), Expect = 0.091
Identities = 19/43 (44%), Positives = 26/43 (60%)
Frame = +1
Query: 66 GKSLGVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSF 108
GK++G+VGLG +G K AFG V+ S S+K EA S+
Sbjct: 310 GKAVGIVGLGRIGSAIAKRAAAFGCPVSYH--SRSEKPEAGSY 432
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.136 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,313,217
Number of Sequences: 36976
Number of extensions: 68843
Number of successful extensions: 356
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 355
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 355
length of query: 194
length of database: 9,014,727
effective HSP length: 91
effective length of query: 103
effective length of database: 5,649,911
effective search space: 581940833
effective search space used: 581940833
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC123898.9


