

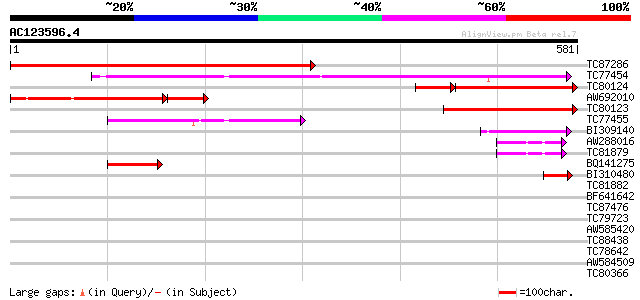
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123596.4 - phase: 0
(581 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87286 similar to GP|22335707|dbj|BAC10552. nine-cis-epoxycarot... 620 e-178
TC77454 homologue to GP|22335695|dbj|BAC10549. nine-cis-epoxycar... 350 9e-97
TC80124 homologue to GP|22335707|dbj|BAC10552. nine-cis-epoxycar... 258 8e-87
AW692010 similar to GP|22335707|db nine-cis-epoxycarotenoid diox... 230 4e-79
TC80123 similar to GP|22335707|dbj|BAC10552. nine-cis-epoxycarot... 259 2e-69
TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarot... 159 2e-39
BI309140 similar to GP|22335695|db nine-cis-epoxycarotenoid diox... 73 3e-13
AW288016 similar to GP|21902049|db hypothetical protein~similar ... 61 1e-09
TC81879 similar to GP|21902049|dbj|BAC05598. hypothetical protei... 61 1e-09
BQ141275 similar to GP|22335695|dbj nine-cis-epoxycarotenoid dio... 60 2e-09
BI310480 homologue to GP|22335703|dbj nine-cis-epoxycarotenoid d... 42 7e-04
TC81882 similar to GP|9759349|dbj|BAB10004.1 contains similarity... 37 0.021
BF641642 weakly similar to GP|16604595|gb| unknown protein {Arab... 36 0.047
TC87476 homologue to PIR|T11622|T11622 extensin class 1 precurso... 35 0.061
TC79723 homologue to SP|Q01520|EF1A_PODAN Elongation factor 1-al... 35 0.10
AW585420 weakly similar to GP|21902049|dbj hypothetical protein~... 35 0.10
TC88438 weakly similar to PIR|E84614|E84614 probable RNA-binding... 34 0.14
TC78642 similar to GP|21553735|gb|AAM62828.1 unknown {Arabidopsi... 34 0.18
AW584509 homologue to GP|13646986|db DNA-binding protein DF1 {Pi... 34 0.18
TC80366 similar to GP|13877617|gb|AAK43886.1 protein kinase-like... 33 0.23
>TC87286 similar to GP|22335707|dbj|BAC10552. nine-cis-epoxycarotenoid
dioxygenase4 {Pisum sativum}, partial (50%)
Length = 987
Score = 620 bits (1599), Expect = e-178
Identities = 306/314 (97%), Positives = 306/314 (97%), Gaps = 1/314 (0%)
Frame = +1
Query: 1 MVPKPIIITSKQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTIT 60
MVPKPIIITSKQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTIT
Sbjct: 46 MVPKPIIITSKQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTIT 225
Query: 61 TTHNLKKPQTSSLPALLFNTFDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPTQCK 120
TTHNLKKPQTSSLPALLFNTFDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPTQCK
Sbjct: 226 TTHNLKKPQTSSLPALLFNTFDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPTQCK 405
Query: 121 VIKGTLPPSLNGVYIRNGPNPQFLPRGPYHLFDGDGMLHAITISNGKATLCSRYVQTYKY 180
VIKGTLPPSLNGVYIRNGPNPQFLPRGPYHLFDGDGMLHAITISNGKATLCSRYVQTYKY
Sbjct: 406 VIKGTLPPSLNGVYIRNGPNPQFLPRGPYHLFDGDGMLHAITISNGKATLCSRYVQTYKY 585
Query: 181 KIENEAGYQLIPNVFSGFNSLIASAARGSVTAARVISGQYNPSNGIGLANTSLALFGNKL 240
KIENEAGYQLIPNVFSGFNSLIASAARGSVTAARVISGQYNPSNGIGLANTSLALFGNKL
Sbjct: 586 KIENEAGYQLIPNVFSGFNSLIASAARGSVTAARVISGQYNPSNGIGLANTSLALFGNKL 765
Query: 241 FALGESDLPYEINLTPNGDIQTIGRYDFNGKLSMSMTAHPKIDGDTGETFAFRYGPMPPF 300
FALGESDLPYEINLTPNGDIQTIGRYDFNGKLSMSMTAHPKIDGDTGETFAFRYGPMPPF
Sbjct: 766 FALGESDLPYEINLTPNGDIQTIGRYDFNGKLSMSMTAHPKIDGDTGETFAFRYGPMPPF 945
Query: 301 LTYFR-FDANGVKH 313
F NGVKH
Sbjct: 946 FNLFSVLMPNGVKH 987
>TC77454 homologue to GP|22335695|dbj|BAC10549. nine-cis-epoxycarotenoid
dioxygenase1 {Pisum sativum}, partial (98%)
Length = 1959
Score = 350 bits (898), Expect = 9e-97
Identities = 200/512 (39%), Positives = 290/512 (56%), Gaps = 20/512 (3%)
Frame = +2
Query: 84 IINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQF 143
I+ F D + P H L+ NFAPV DE PP + IKG LP LNG ++R GPN +F
Sbjct: 176 IVKLFYDSSL-----PHHWLAGNFAPVKDETPPIKDLPIKGHLPDCLNGEFVRVGPNSKF 340
Query: 144 LPRGPYHLFDGDGMLHAITISNGKATLCSRYVQTYKYKIENEAGYQLIPNV--FSGFNSL 201
P YH FDGDGM+H + I +GKAT SR+V+T + K E G + G L
Sbjct: 341 APVAGYHWFDGDGMIHGLRIKDGKATYVSRFVKTSRLKQEEYFGGSKFMKIGDMKGLFGL 520
Query: 202 IASAARGSVTAARVISGQYNPSNGIGLANTSLALFGNKLFALGESDLPYEINLTPNGDIQ 261
+ + +V+ Y G G ANT+L KL AL E D PY I + +GD+Q
Sbjct: 521 LMVNMQMLRAKLKVVDVSY----GHGTANTALVYHHQKLLALSEGDKPYAIKVFEDGDLQ 688
Query: 262 TIGRYDFNGKLSMSMTAHPKIDGDTGETFAFRYGPMPPFLTYFRFDANGVKHNDVPVFSM 321
T+G D++ +L + TAHPK+D TGE F F Y PP++TY +G H+ VP+ ++
Sbjct: 689 TLGMLDYDKRLGHNFTAHPKVDPFTGEMFTFGYSHTPPYITYRVISKDGFMHDPVPI-TI 865
Query: 322 TRPSFLHDFAITKKYAVFTDIQLGMNPLDMISGGSPVGS-DPSKVSRIGILPRYASDESK 380
+ P +HDFAIT+ Y++F D+ L P +M+ + + S D +K +R G+LPRYA DE
Sbjct: 866 SEPIMMHDFAITENYSIFMDLPLYFRPKEMVKNKTLIFSFDSTKKARFGVLPRYAKDEKH 1045
Query: 381 MKWFDVPGFNIMHAINSWDGEDDEETVT--LIAPNVLSIEHTM-ERLELVHAMIEKVKIN 437
++WF++P I H N+W+ ED+ +T L PN+ + + E+L+ + +++ N
Sbjct: 1046IRWFELPNCFIFHNANAWEEEDEIVLITCRLENPNLDMVGGAVKEKLDNFANELYEMRFN 1225
Query: 438 IKTGIVSRQPLSARNLDFAVINGDYMGKKNRFVYAAIGNPMPKISGVVKIDV-------- 489
+KTG S++ LSA +DF +N Y G+K R+VY + + K++G+VK D+
Sbjct: 1226MKTGEASQKKLSASAVDFPRVNESYTGRKQRYVYGTELDSIAKVTGIVKFDLHAKPDSGK 1405
Query: 490 ----LKGEEVGCRLYGEGCYGGEPFFVAREDG--EEEDDGYLVSYVHDEKKGESKFLVMD 543
+ G VG G G +G E +V R G EEDDGYL+ +VHDE G+S V+D
Sbjct: 1406TKLEVGGNVVGHYDLGPGRFGSEAVYVPRVPGTDSEEDDGYLIFFVHDENTGKSFVHVLD 1585
Query: 544 AKSPEFEIVAEVKLPRRVPYGFHGLFVKESDI 575
AK+ + VA V+LP+RVPYGFH FV E +
Sbjct: 1586AKTMSADPVAVVELPQRVPYGFHAFFVTEEQL 1681
>TC80124 homologue to GP|22335707|dbj|BAC10552. nine-cis-epoxycarotenoid
dioxygenase4 {Pisum sativum}, partial (28%)
Length = 758
Score = 258 bits (658), Expect(2) = 8e-87
Identities = 125/125 (100%), Positives = 125/125 (100%)
Frame = +3
Query: 457 VINGDYMGKKNRFVYAAIGNPMPKISGVVKIDVLKGEEVGCRLYGEGCYGGEPFFVARED 516
VINGDYMGKKNRFVYAAIGNPMPKISGVVKIDVLKGEEVGCRLYGEGCYGGEPFFVARED
Sbjct: 126 VINGDYMGKKNRFVYAAIGNPMPKISGVVKIDVLKGEEVGCRLYGEGCYGGEPFFVARED 305
Query: 517 GEEEDDGYLVSYVHDEKKGESKFLVMDAKSPEFEIVAEVKLPRRVPYGFHGLFVKESDIT 576
GEEEDDGYLVSYVHDEKKGESKFLVMDAKSPEFEIVAEVKLPRRVPYGFHGLFVKESDIT
Sbjct: 306 GEEEDDGYLVSYVHDEKKGESKFLVMDAKSPEFEIVAEVKLPRRVPYGFHGLFVKESDIT 485
Query: 577 KLSVS 581
KLSVS
Sbjct: 486 KLSVS 500
Score = 81.6 bits (200), Expect(2) = 8e-87
Identities = 41/41 (100%), Positives = 41/41 (100%)
Frame = +2
Query: 416 SIEHTMERLELVHAMIEKVKINIKTGIVSRQPLSARNLDFA 456
SIEHTMERLELVHAMIEKVKINIKTGIVSRQPLSARNLDFA
Sbjct: 2 SIEHTMERLELVHAMIEKVKINIKTGIVSRQPLSARNLDFA 124
>AW692010 similar to GP|22335707|db nine-cis-epoxycarotenoid dioxygenase4
{Pisum sativum}, partial (21%)
Length = 653
Score = 230 bits (587), Expect(2) = 4e-79
Identities = 119/162 (73%), Positives = 130/162 (79%), Gaps = 1/162 (0%)
Frame = +2
Query: 1 MVPKPIII-TSKQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTI 59
MVPKPIII T+KQPPT IP FNI+ ++T+EKPQTSQ T + TKTL T
Sbjct: 26 MVPKPIIILTTKQPPTIP-IPQFNIAFVRTQEKPQTSQPHETLRTGSPAKTKTLLTSQKH 202
Query: 60 TTTHNLKKPQTSSLPALLFNTFDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPTQC 119
T+ K+ QTSS+ AL+ NT DDIINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPTQC
Sbjct: 203 KNTN--KRLQTSSISALILNTLDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPTQC 376
Query: 120 KVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHLFDGDGMLHAI 161
+IKGTLPPSL G YIRNGPNPQFLPRGPYHLFDGDGMLHAI
Sbjct: 377 VIIKGTLPPSLYGAYIRNGPNPQFLPRGPYHLFDGDGMLHAI 502
Score = 83.2 bits (204), Expect(2) = 4e-79
Identities = 39/42 (92%), Positives = 42/42 (99%)
Frame = +3
Query: 162 TISNGKATLCSRYVQTYKYKIENEAGYQLIPNVFSGFNSLIA 203
TISNGKATLCSRYV+TYKYKIEN+AG+QLIPNVFSGFNSLIA
Sbjct: 504 TISNGKATLCSRYVKTYKYKIENKAGHQLIPNVFSGFNSLIA 629
>TC80123 similar to GP|22335707|dbj|BAC10552. nine-cis-epoxycarotenoid
dioxygenase4 {Pisum sativum}, partial (23%)
Length = 657
Score = 259 bits (663), Expect = 2e-69
Identities = 126/137 (91%), Positives = 130/137 (93%)
Frame = +3
Query: 445 RQPLSARNLDFAVINGDYMGKKNRFVYAAIGNPMPKISGVVKIDVLKGEEVGCRLYGEGC 504
R PLSARNLD AVI D++GKK+RFVYAAIGNPMPK SGVVKIDVLKGEEVGCRLYGEGC
Sbjct: 3 RHPLSARNLDLAVITNDFLGKKSRFVYAAIGNPMPKFSGVVKIDVLKGEEVGCRLYGEGC 182
Query: 505 YGGEPFFVAREDGEEEDDGYLVSYVHDEKKGESKFLVMDAKSPEFEIVAEVKLPRRVPYG 564
YGGEPFFVA E GEEEDDGYLVSYVHDEKKGESKFLVMDAKSPEFEIVAEVKLPRRVPYG
Sbjct: 183 YGGEPFFVAMEGGEEEDDGYLVSYVHDEKKGESKFLVMDAKSPEFEIVAEVKLPRRVPYG 362
Query: 565 FHGLFVKESDITKLSVS 581
FHGLFVK+SDITKLSVS
Sbjct: 363 FHGLFVKQSDITKLSVS 413
>TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarotenoid
dioxygenase1 {Pisum sativum}, partial (43%)
Length = 1865
Score = 159 bits (403), Expect = 2e-39
Identities = 87/206 (42%), Positives = 115/206 (55%), Gaps = 3/206 (1%)
Frame = +3
Query: 101 HVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHLFDGDGMLHA 160
H L+ NFAPV DE PP + +KG LP LNG ++R GPNP+F P YH FDGDGM+H
Sbjct: 1251 HWLAGNFAPVQDETPPIKDLPVKGHLPDCLNGEFVRVGPNPKFSPVAGYHWFDGDGMIHG 1430
Query: 161 ITISNGKATLCSRYVQTYKYKIENEAG---YQLIPNVFSGFNSLIASAARGSVTAARVIS 217
+ I +GKAT S +V+T + K E G + I +V G L+ + +++
Sbjct: 1431 LRIKDGKATYVSHFVRTSRLKQEEYFGGCKFMKIGDV-KGLLGLLMVTIQTLREKLKILD 1607
Query: 218 GQYNPSNGIGLANTSLALFGNKLFALGESDLPYEINLTPNGDIQTIGRYDFNGKLSMSMT 277
Y G G ANT L KL AL E D PY I + +GD+ T+G D+ +L T
Sbjct: 1608 VSY----GTGTANTGLVYHNGKLLALSERDKPYAIKVFEDGDLHTLGMLDYEKRLDHYFT 1775
Query: 278 AHPKIDGDTGETFAFRYGPMPPFLTY 303
AHPK+D TGE F F Y PP++TY
Sbjct: 1776 AHPKVDPFTGEMFTFGYSQTPPYITY 1853
>BI309140 similar to GP|22335695|db nine-cis-epoxycarotenoid dioxygenase1
{Pisum sativum}, partial (18%)
Length = 589
Score = 72.8 bits (177), Expect = 3e-13
Identities = 39/95 (41%), Positives = 57/95 (59%), Gaps = 2/95 (2%)
Frame = +3
Query: 483 GVVKIDVLKGEEVGCRLYGEGCYGGEPFFVAREDG--EEEDDGYLVSYVHDEKKGESKFL 540
G K+++ G G G+G + +P +V R G +EDDGYL+ +VHDE +S
Sbjct: 12 GKTKLEI-GGNVHGIYDLGQGTFCSDPIYVPRVPGTDSDEDDGYLIFFVHDENTRKSFVH 188
Query: 541 VMDAKSPEFEIVAEVKLPRRVPYGFHGLFVKESDI 575
V+DAK+ + VA V+LP+RVPYGFH F+ E +
Sbjct: 189 VIDAKTMSADPVAVVELPQRVPYGFHSFFMTEDQL 293
>AW288016 similar to GP|21902049|db hypothetical protein~similar to
Arabidopsis thaliana chromosome 4 At4g32810, partial
(17%)
Length = 542
Score = 60.8 bits (146), Expect = 1e-09
Identities = 32/71 (45%), Positives = 41/71 (57%)
Frame = +3
Query: 500 YGEGCYGGEPFFVAREDGEEEDDGYLVSYVHDEKKGESKFLVMDAKSPEFEIVAEVKLPR 559
Y EG EPFFV R +EDDG ++S + EK GE LV+D + FE +A K P
Sbjct: 99 YEEGAVPSEPFFVPRPGATKEDDGVVISII-SEKNGEGYALVLDGST--FEEIARAKFPY 269
Query: 560 RVPYGFHGLFV 570
+PYG HG +V
Sbjct: 270 GLPYGLHGCWV 302
>TC81879 similar to GP|21902049|dbj|BAC05598. hypothetical protein~similar
to Arabidopsis thaliana chromosome 4 At4g32810, partial
(15%)
Length = 520
Score = 60.8 bits (146), Expect = 1e-09
Identities = 32/71 (45%), Positives = 41/71 (57%)
Frame = +3
Query: 500 YGEGCYGGEPFFVAREDGEEEDDGYLVSYVHDEKKGESKFLVMDAKSPEFEIVAEVKLPR 559
Y EG EPFFV R +EDDG ++S + EK GE LV+D + FE +A K P
Sbjct: 177 YEEGAVPSEPFFVPRPGATKEDDGVVISII-SEKNGEGYALVLDGST--FEEIARAKFPY 347
Query: 560 RVPYGFHGLFV 570
+PYG HG +V
Sbjct: 348 GLPYGLHGCWV 380
>BQ141275 similar to GP|22335695|dbj nine-cis-epoxycarotenoid dioxygenase1
{Pisum sativum}, partial (17%)
Length = 595
Score = 60.5 bits (145), Expect = 2e-09
Identities = 28/56 (50%), Positives = 35/56 (62%)
Frame = +2
Query: 101 HVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHLFDGDG 156
H ++ NFAPV DE PPT+ +KG LP LNG ++R GPNP+ P Y DG G
Sbjct: 428 HWIAGNFAPVKDETPPTKDLPVKGHLPDCLNGEFVRVGPNPKXXPVAGYXWLDGXG 595
>BI310480 homologue to GP|22335703|dbj nine-cis-epoxycarotenoid dioxygenase3
{Pisum sativum}, partial (5%)
Length = 415
Score = 42.0 bits (97), Expect = 7e-04
Identities = 17/29 (58%), Positives = 22/29 (75%)
Frame = +3
Query: 548 EFEIVAEVKLPRRVPYGFHGLFVKESDIT 576
+ ++ A VKLP RVPYGFHG FV+ D+T
Sbjct: 12 DLKVEATVKLPSRVPYGFHGTFVEAKDLT 98
>TC81882 similar to GP|9759349|dbj|BAB10004.1 contains similarity to unknown
protein~gb|AAF72944.1~gene_id:MAH20.11 {Arabidopsis
thaliana}, partial (5%)
Length = 875
Score = 37.0 bits (84), Expect = 0.021
Identities = 23/64 (35%), Positives = 29/64 (44%)
Frame = +3
Query: 9 TSKQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTITTTHNLKKP 68
T+ PP+ + PP N+ K P S S T K+T T P P PT T + L P
Sbjct: 168 TTLLPPSLPNPPPLNL-----KNPPNFSVSPTTKSTPITKPLVHAPPNPTTTAPNPLLPP 332
Query: 69 QTSS 72
T S
Sbjct: 333 LTKS 344
>BF641642 weakly similar to GP|16604595|gb| unknown protein {Arabidopsis
thaliana}, partial (12%)
Length = 563
Score = 35.8 bits (81), Expect = 0.047
Identities = 29/72 (40%), Positives = 36/72 (49%)
Frame = +3
Query: 9 TSKQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTITTTHNLKKP 68
TS QPP+ S+ F S + S S TTTTTPPT T PT TT ++
Sbjct: 54 TSPQPPSLSNALHFFTSLLNR----YASVSTPSPTTTTTPPTTTTPTG---TTAGGIRCS 212
Query: 69 QTSSLPALLFNT 80
+ SS PA L N+
Sbjct: 213 RNSS-PAQLNNS 245
>TC87476 homologue to PIR|T11622|T11622 extensin class 1 precursor - cowpea,
partial (48%)
Length = 939
Score = 35.4 bits (80), Expect = 0.061
Identities = 38/146 (26%), Positives = 51/146 (34%)
Frame = +2
Query: 3 PKPIIITSKQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTITTT 62
P P I S PP+ S PP+ K P S S +PP + P+PP
Sbjct: 2 PPPYIYKSPPPPSPSPPPPY-----VYKSPPPPSPSPPPPYVYKSPPPPS-PSPPPPYVY 163
Query: 63 HNLKKPQTSSLPALLFNTFDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPTQCKVI 122
+ P S P ++ + PP PS P +V P PP V
Sbjct: 164 KSPPPPTPSPPPPYIYKS---------PPPPSPSPPPPYVYKSPPPPSPSPPPP---YVY 307
Query: 123 KGTLPPSLNGVYIRNGPNPQFLPRGP 148
K PPS P P ++ + P
Sbjct: 308 KSPPPPS-------PSPPPPYIYKSP 364
Score = 30.8 bits (68), Expect = 1.5
Identities = 31/129 (24%), Positives = 44/129 (34%), Gaps = 6/129 (4%)
Frame = +2
Query: 3 PKPIIITSKQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTITTT 62
P P + S PPT S PP+ K P S S +PP + P+PP
Sbjct: 146 PPPYVYKSPPPPTPSPPPPYIY-----KSPPPPSPSPPPPYVYKSPPPPS-PSPPPPYVY 307
Query: 63 HNLKKPQTSSLPALLFNT------FDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDELPP 116
+ P S P ++ + F P S P + Q +P + P
Sbjct: 308 KSPPPPSPSPPPPYIYKSPPTTIPFTTTSICL*VPSSSFSFTPSSICLQISSPTISFTPS 487
Query: 117 TQCKVIKGT 125
T C + T
Sbjct: 488 TLCL*VTST 514
Score = 30.0 bits (66), Expect = 2.6
Identities = 38/173 (21%), Positives = 55/173 (30%), Gaps = 4/173 (2%)
Frame = +3
Query: 3 PKPIIITSKQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTITTT 62
P P + S PP+ S PP+ S PP P PP + +
Sbjct: 387 PPPYVYKSPPPPSPSPPPPYVYKS--------------------PPPPSPSPPPPYVYKS 506
Query: 63 HNLKKPQTSSLPALLFNTFDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPTQCKVI 122
P S P + + PP PS P + P PP +
Sbjct: 507 P--PPPSASPPPPYYYKS---------PPPPSPSPPPPYGYKSPPPPSPSPPPP---YIY 644
Query: 123 KGTLPPSLN----GVYIRNGPNPQFLPRGPYHLFDGDGMLHAITISNGKATLC 171
K PPS + Y+ N P P Y+L ++ +I + N + C
Sbjct: 645 KSPPPPSPSPPPYHPYLYNSPPPPIY*ASGYYLKKFQ*IIFSIVVVNLYSIFC 803
>TC79723 homologue to SP|Q01520|EF1A_PODAN Elongation factor 1-alpha
(EF-1-alpha). {Podospora anserina}, complete
Length = 1537
Score = 34.7 bits (78), Expect = 0.10
Identities = 29/107 (27%), Positives = 47/107 (43%), Gaps = 10/107 (9%)
Frame = +2
Query: 3 PKPIIITSKQPPTTSSIPPFNI--SSIKTKEKPQTSQSQTLKTTTTTP--------PTKT 52
P P + +S P+T PP + ++ ++ +P TS ++ T P T
Sbjct: 497 PTPWVSSSSSSPSTRWTPPSGLRTATRRSSRRPPTSSRKSATTPRLLPSSPSPVSTATTC 676
Query: 53 LPTPPTITTTHNLKKPQTSSLPALLFNTFDDIINTFIDPPIKPSVDP 99
LP PPT+ T KK S + T + I+ I+PP +P+ P
Sbjct: 677 LPPPPTLLGTRVGKKEAKSG--KVTGKTLLEAIDA-IEPPKRPTDKP 808
>AW585420 weakly similar to GP|21902049|dbj hypothetical protein~similar to
Arabidopsis thaliana chromosome 4 At4g32810, partial
(13%)
Length = 551
Score = 34.7 bits (78), Expect = 0.10
Identities = 20/64 (31%), Positives = 33/64 (51%), Gaps = 3/64 (4%)
Frame = +3
Query: 121 VIKGTLPPSLNGVYIRNGPNPQFLPRGPY---HLFDGDGMLHAITISNGKATLCSRYVQT 177
+++G +P L G Y++NGP + G Y HLFDG L + +G+ R V++
Sbjct: 357 LVQGHIPLWLKGTYLKNGPGMWNI--GDYNFRHLFDGYATLVGLHFEDGRLIAGHRQVES 530
Query: 178 YKYK 181
Y+
Sbjct: 531 QAYQ 542
>TC88438 weakly similar to PIR|E84614|E84614 probable RNA-binding protein
[imported] - Arabidopsis thaliana, partial (9%)
Length = 1622
Score = 34.3 bits (77), Expect = 0.14
Identities = 23/59 (38%), Positives = 31/59 (51%), Gaps = 5/59 (8%)
Frame = +3
Query: 18 SIPPFNISSIKTKEKPQTSQS-----QTLKTTTTTPPTKTLPTPPTITTTHNLKKPQTS 71
S P N++ IKT+ Q SQ+ Q KT + PP K P ITTTHN + +T+
Sbjct: 126 SSQPINVNFIKTQNPFQKSQTFYNFVQKCKTPLSPPPLK-----PPITTTHNHR*NETT 287
>TC78642 similar to GP|21553735|gb|AAM62828.1 unknown {Arabidopsis
thaliana}, partial (90%)
Length = 691
Score = 33.9 bits (76), Expect = 0.18
Identities = 14/35 (40%), Positives = 21/35 (60%)
Frame = +1
Query: 25 SSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTI 59
S I T P +S++ TL TT+ +PP+ P+P I
Sbjct: 106 SLIATHHSPPSSETSTLSTTSVSPPSPAFPSPSVI 210
>AW584509 homologue to GP|13646986|db DNA-binding protein DF1 {Pisum
sativum}, partial (22%)
Length = 694
Score = 33.9 bits (76), Expect = 0.18
Identities = 26/87 (29%), Positives = 38/87 (42%)
Frame = +2
Query: 30 KEKPQTSQSQTLKTTTTTPPTKTLPTPPTITTTHNLKKPQTSSLPALLFNTFDDIINTFI 89
+ P QS ++ T PP T+PTP TT L P T+++P+ N
Sbjct: 452 ENNPSMHQSPSI----TPPPISTIPTP--TTTPSFLSAPPTTTVPSTTIPMSYTQSNPTH 613
Query: 90 DPPIKPSVDPRHVLSQNFAPVLDELPP 116
PPI S +P + + N P + PP
Sbjct: 614 FPPIPSSTNPINTI--NPIPQITTTPP 688
>TC80366 similar to GP|13877617|gb|AAK43886.1 protein kinase-like protein
{Arabidopsis thaliana}, partial (16%)
Length = 814
Score = 33.5 bits (75), Expect = 0.23
Identities = 18/62 (29%), Positives = 27/62 (43%)
Frame = +3
Query: 10 SKQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTITTTHNLKKPQ 69
S PP T S PP + + P T T ++ PP+ T PP T +++ P
Sbjct: 3 SSPPPATPSSPPPSTPANPPPVTPSAPPPSTPATPSSPPPSTTPSAPPPSTPSNSPPSPP 182
Query: 70 TS 71
T+
Sbjct: 183TT 188
Score = 32.3 bits (72), Expect = 0.52
Identities = 21/65 (32%), Positives = 27/65 (41%), Gaps = 1/65 (1%)
Frame = +3
Query: 5 PIIITSKQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKT-LPTPPTITTTH 63
P S PP+T + PP S P T S TT + PP T +PP+ TT
Sbjct: 12 PPATPSSPPPSTPANPPPVTPSAPPPSTPATPSSPPPSTTPSAPPPSTPSNSPPSPPTTP 191
Query: 64 NLKKP 68
+ P
Sbjct: 192AISPP 206
Score = 29.6 bits (65), Expect = 3.4
Identities = 23/77 (29%), Positives = 32/77 (40%), Gaps = 8/77 (10%)
Frame = +3
Query: 5 PIIITSKQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKT--TTTTPPTKT------LPTP 56
P +S P TT S PP + S P T TT +PP++T P+P
Sbjct: 96 PATPSSPPPSTTPSAPPPSTPSNSPPSPPTTPAISPPSGGGTTPSPPSRTPPSSDDSPSP 275
Query: 57 PTITTTHNLKKPQTSSL 73
P+ T P +SS+
Sbjct: 276 PSSKTPPPPSPPSSSSI 326
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.137 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,385,156
Number of Sequences: 36976
Number of extensions: 279302
Number of successful extensions: 2529
Number of sequences better than 10.0: 171
Number of HSP's better than 10.0 without gapping: 2170
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2375
length of query: 581
length of database: 9,014,727
effective HSP length: 101
effective length of query: 480
effective length of database: 5,280,151
effective search space: 2534472480
effective search space used: 2534472480
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC123596.4


