

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123596.12 - phase: 0
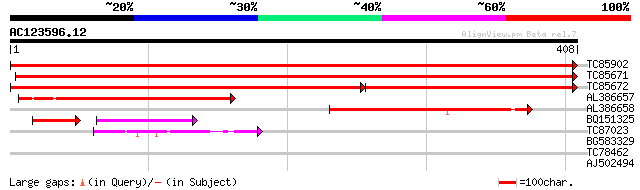
(408 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85902 similar to SP|Q9XF97|RL4_PRUAR 60S ribosomal protein L4 ... 802 0.0
TC85671 similar to SP|Q9XF97|RL4_PRUAR 60S ribosomal protein L4 ... 756 0.0
TC85672 similar to SP|Q9XF97|RL4_PRUAR 60S ribosomal protein L4 ... 480 0.0
AL386657 similar to GP|7630164|emb 60s ribosomal protein l2 {Sch... 224 5e-59
AL386658 weakly similar to PIR|A24579|R5XL ribosomal protein XL1... 142 2e-34
BQ151325 61 1e-18
TC87023 similar to SP|O80361|RK4_TOBAC 50S ribosomal protein L4 ... 47 1e-05
BG583329 similar to PIR|T00956|T00 translation initiation factor... 30 1.7
TC78462 similar to GP|21592780|gb|AAM64729.1 nucleic acid bindin... 29 3.8
AJ502494 similar to GP|7547109|gb| unknown protein {Arabidopsis ... 28 8.4
>TC85902 similar to SP|Q9XF97|RL4_PRUAR 60S ribosomal protein L4 (L1).
[Apricot] {Prunus armeniaca}, complete
Length = 1571
Score = 802 bits (2071), Expect = 0.0
Identities = 408/408 (100%), Positives = 408/408 (100%)
Frame = +1
Query: 1 MAAAAAAARPLVTIQTLESEMSTDSPNTLPIPDVMRASIRPDIVNFVHSNISKNSRQPYA 60
MAAAAAAARPLVTIQTLESEMSTDSPNTLPIPDVMRASIRPDIVNFVHSNISKNSRQPYA
Sbjct: 73 MAAAAAAARPLVTIQTLESEMSTDSPNTLPIPDVMRASIRPDIVNFVHSNISKNSRQPYA 252
Query: 61 VSRKAGHQTSAESWGTGRAVSRIPRVAGGGTHRAGQAAFGNMCRGGRMFAPTRIWRKWHR 120
VSRKAGHQTSAESWGTGRAVSRIPRVAGGGTHRAGQAAFGNMCRGGRMFAPTRIWRKWHR
Sbjct: 253 VSRKAGHQTSAESWGTGRAVSRIPRVAGGGTHRAGQAAFGNMCRGGRMFAPTRIWRKWHR 432
Query: 121 KINVNQKRYAVVSAIAASAIPSLVLARGHRIETVPEFPLVVGDAAEGVEKTKEAIKVLKQ 180
KINVNQKRYAVVSAIAASAIPSLVLARGHRIETVPEFPLVVGDAAEGVEKTKEAIKVLKQ
Sbjct: 433 KINVNQKRYAVVSAIAASAIPSLVLARGHRIETVPEFPLVVGDAAEGVEKTKEAIKVLKQ 612
Query: 181 VGAFADAEKAKDSLGIRPGKGKMRNRRYISRKGPLIVYGTDGAKAVKAFRNIPGVEITNV 240
VGAFADAEKAKDSLGIRPGKGKMRNRRYISRKGPLIVYGTDGAKAVKAFRNIPGVEITNV
Sbjct: 613 VGAFADAEKAKDSLGIRPGKGKMRNRRYISRKGPLIVYGTDGAKAVKAFRNIPGVEITNV 792
Query: 241 ERLNLLKLAPGGHLGRFVIWTKSAFEKLDSIYGTFDKGSEKKKGYVLPRAKMVNSDLTRI 300
ERLNLLKLAPGGHLGRFVIWTKSAFEKLDSIYGTFDKGSEKKKGYVLPRAKMVNSDLTRI
Sbjct: 793 ERLNLLKLAPGGHLGRFVIWTKSAFEKLDSIYGTFDKGSEKKKGYVLPRAKMVNSDLTRI 972
Query: 301 INSDEVQSVVRPIKKDVKRATLKKNPLKNLNVMLRLNPYAKTAKRMALLAEAERVKSKKE 360
INSDEVQSVVRPIKKDVKRATLKKNPLKNLNVMLRLNPYAKTAKRMALLAEAERVKSKKE
Sbjct: 973 INSDEVQSVVRPIKKDVKRATLKKNPLKNLNVMLRLNPYAKTAKRMALLAEAERVKSKKE 1152
Query: 361 KFDKKRKIVSKEEASAIKAAGKAWYNTMVSDSDYAEFDNFSKWLGVSQ 408
KFDKKRKIVSKEEASAIKAAGKAWYNTMVSDSDYAEFDNFSKWLGVSQ
Sbjct: 1153KFDKKRKIVSKEEASAIKAAGKAWYNTMVSDSDYAEFDNFSKWLGVSQ 1296
>TC85671 similar to SP|Q9XF97|RL4_PRUAR 60S ribosomal protein L4 (L1).
[Apricot] {Prunus armeniaca}, complete
Length = 1508
Score = 756 bits (1951), Expect = 0.0
Identities = 376/404 (93%), Positives = 394/404 (97%)
Frame = +3
Query: 5 AAAARPLVTIQTLESEMSTDSPNTLPIPDVMRASIRPDIVNFVHSNISKNSRQPYAVSRK 64
AAAARPLVT+ T+E +M+TD+P +LP+PDVMRA+IRPDIVNFVHSNISKN+RQPYAVSRK
Sbjct: 60 AAAARPLVTVHTVEGDMATDTPTSLPVPDVMRAAIRPDIVNFVHSNISKNARQPYAVSRK 239
Query: 65 AGHQTSAESWGTGRAVSRIPRVAGGGTHRAGQAAFGNMCRGGRMFAPTRIWRKWHRKINV 124
AGHQTSAESWGTGRAVSRIPRVAGGGTHRAGQAAFGNMCRGGRMFAPTRIWRKWHRKINV
Sbjct: 240 AGHQTSAESWGTGRAVSRIPRVAGGGTHRAGQAAFGNMCRGGRMFAPTRIWRKWHRKINV 419
Query: 125 NQKRYAVVSAIAASAIPSLVLARGHRIETVPEFPLVVGDAAEGVEKTKEAIKVLKQVGAF 184
NQKRYAVVSAIAASAIPSLVLARGHRIETVPE PLVVGD+AEGVEKTKEAIK+LK +GA+
Sbjct: 420 NQKRYAVVSAIAASAIPSLVLARGHRIETVPELPLVVGDSAEGVEKTKEAIKLLKSIGAY 599
Query: 185 ADAEKAKDSLGIRPGKGKMRNRRYISRKGPLIVYGTDGAKAVKAFRNIPGVEITNVERLN 244
DAEKAKDSLGIRPGKGKMRNRRYISRKGPLIVYGT+GAKAVKAFRNIPGVEITNVERLN
Sbjct: 600 PDAEKAKDSLGIRPGKGKMRNRRYISRKGPLIVYGTEGAKAVKAFRNIPGVEITNVERLN 779
Query: 245 LLKLAPGGHLGRFVIWTKSAFEKLDSIYGTFDKGSEKKKGYVLPRAKMVNSDLTRIINSD 304
LLKLAPGGHLGRFVIWTK+AFEKLDSIYG+FDK SEKKKGYVLPRAKMVNSDLTRIINSD
Sbjct: 780 LLKLAPGGHLGRFVIWTKTAFEKLDSIYGSFDKPSEKKKGYVLPRAKMVNSDLTRIINSD 959
Query: 305 EVQSVVRPIKKDVKRATLKKNPLKNLNVMLRLNPYAKTAKRMALLAEAERVKSKKEKFDK 364
EVQSVVRP+KKDVKRATLKKNPLKNLNVMLRLNPYAKTAKRMALLAEAERVKSKKEK DK
Sbjct: 960 EVQSVVRPVKKDVKRATLKKNPLKNLNVMLRLNPYAKTAKRMALLAEAERVKSKKEKLDK 1139
Query: 365 KRKIVSKEEASAIKAAGKAWYNTMVSDSDYAEFDNFSKWLGVSQ 408
KRK VSKEEA+AIKAAGKAWYNTMVSDSDY EFDNFSKWLGVSQ
Sbjct: 1140KRKTVSKEEATAIKAAGKAWYNTMVSDSDYTEFDNFSKWLGVSQ 1271
>TC85672 similar to SP|Q9XF97|RL4_PRUAR 60S ribosomal protein L4 (L1).
[Apricot] {Prunus armeniaca}, complete
Length = 1507
Score = 480 bits (1235), Expect(2) = 0.0
Identities = 239/256 (93%), Positives = 249/256 (96%)
Frame = +1
Query: 1 MAAAAAAARPLVTIQTLESEMSTDSPNTLPIPDVMRASIRPDIVNFVHSNISKNSRQPYA 60
+ A+AAARPLVT+QTL+S+M+TDS TLPIPDVMRASIRPDIVNFVHSNISKNSRQPYA
Sbjct: 7 LTMASAAARPLVTVQTLDSDMATDSQTTLPIPDVMRASIRPDIVNFVHSNISKNSRQPYA 186
Query: 61 VSRKAGHQTSAESWGTGRAVSRIPRVAGGGTHRAGQAAFGNMCRGGRMFAPTRIWRKWHR 120
VSRKAGHQTSAESWGTGRAVSRIPRVAGGGTHRAGQAAFGNMCRGGRMFAPTRIWRKWHR
Sbjct: 187 VSRKAGHQTSAESWGTGRAVSRIPRVAGGGTHRAGQAAFGNMCRGGRMFAPTRIWRKWHR 366
Query: 121 KINVNQKRYAVVSAIAASAIPSLVLARGHRIETVPEFPLVVGDAAEGVEKTKEAIKVLKQ 180
KINVNQKRYAVVSAIAASAIPSLVLARGHRIETVPEFPLVVGD+AEGVEKTKEAI VLK+
Sbjct: 367 KINVNQKRYAVVSAIAASAIPSLVLARGHRIETVPEFPLVVGDSAEGVEKTKEAISVLKK 546
Query: 181 VGAFADAEKAKDSLGIRPGKGKMRNRRYISRKGPLIVYGTDGAKAVKAFRNIPGVEITNV 240
+GAFADAEKAK SLGIRPGKGKMRNRRYISRKGPLIVYGT+GAK VKAFRNIPGVEITNV
Sbjct: 547 IGAFADAEKAKASLGIRPGKGKMRNRRYISRKGPLIVYGTEGAKLVKAFRNIPGVEITNV 726
Query: 241 ERLNLLKLAPGGHLGR 256
ERLNLLKLAPGGHLGR
Sbjct: 727 ERLNLLKLAPGGHLGR 774
Score = 292 bits (748), Expect(2) = 0.0
Identities = 147/152 (96%), Positives = 149/152 (97%)
Frame = +2
Query: 257 FVIWTKSAFEKLDSIYGTFDKGSEKKKGYVLPRAKMVNSDLTRIINSDEVQSVVRPIKKD 316
FV+WTKSAFEKLDSIYGTFDKGSEKKKGYVLPRAKMVNSDLTRIINSDEVQSVVRPIKKD
Sbjct: 776 FVVWTKSAFEKLDSIYGTFDKGSEKKKGYVLPRAKMVNSDLTRIINSDEVQSVVRPIKKD 955
Query: 317 VKRATLKKNPLKNLNVMLRLNPYAKTAKRMALLAEAERVKSKKEKFDKKRKIVSKEEASA 376
VKRATLKKNPLKNLNVMLRLNPYAKTAKRMALLAEAERVK+KKEK DKKRK VSKEEASA
Sbjct: 956 VKRATLKKNPLKNLNVMLRLNPYAKTAKRMALLAEAERVKAKKEKLDKKRKTVSKEEASA 1135
Query: 377 IKAAGKAWYNTMVSDSDYAEFDNFSKWLGVSQ 408
IKAAGKAWYNTMVSDSDY EFDNFSKWLGVSQ
Sbjct: 1136IKAAGKAWYNTMVSDSDYTEFDNFSKWLGVSQ 1231
>AL386657 similar to GP|7630164|emb 60s ribosomal protein l2
{Schizosaccharomyces pombe}, partial (35%)
Length = 468
Score = 224 bits (571), Expect = 5e-59
Identities = 112/156 (71%), Positives = 129/156 (81%)
Frame = +1
Query: 7 AARPLVTIQTLESEMSTDSPNTLPIPDVMRASIRPDIVNFVHSNISKNSRQPYAVSRKAG 66
A RP + I + ES +T + LP V +A IRPDIV VH+NISK++RQPYAVS KAG
Sbjct: 1 AIRPQINIYS-ESGDNTIGKHVLPA--VFKAPIRPDIVRVVHTNISKSNRQPYAVSSKAG 171
Query: 67 HQTSAESWGTGRAVSRIPRVAGGGTHRAGQAAFGNMCRGGRMFAPTRIWRKWHRKINVNQ 126
HQTSAESWGTGRAV+RIPRV+GGGTHRAGQ AFGNMCRGGRMFAPT+IWRKWH K N+NQ
Sbjct: 172 HQTSAESWGTGRAVARIPRVSGGGTHRAGQGAFGNMCRGGRMFAPTKIWRKWHVKTNLNQ 351
Query: 127 KRYAVVSAIAASAIPSLVLARGHRIETVPEFPLVVG 162
KR+A SA+AAS+IPSLVLARGHRIE + E PLV+G
Sbjct: 352 KRFATASALAASSIPSLVLARGHRIEEIQEVPLVIG 459
>AL386658 weakly similar to PIR|A24579|R5XL ribosomal protein XL1a - African
clawed frog, partial (20%)
Length = 486
Score = 142 bits (358), Expect = 2e-34
Identities = 75/148 (50%), Positives = 100/148 (66%), Gaps = 2/148 (1%)
Frame = +3
Query: 231 NIPGVEITNVERLNLLKLAPGGHLGRFVIWTKSAFEKLDSIYGTFDKGSEKKKGYVLPRA 290
NIPGVE+ NV RLNLL+LAPGGH+GRF+IW++ AF LDS++GT+ K + K+ Y LP
Sbjct: 3 NIPGVELVNVRRLNLLQLAPGGHVGRFIIWSQDAFALLDSLFGTYRKSAALKRDYHLPSH 182
Query: 291 KMVNSDLTRIINSDEVQSVVRPI--KKDVKRATLKKNPLKNLNVMLRLNPYAKTAKRMAL 348
+ NSD+T IINS E+QS++RP K + + T KKNPL+N V +RLNPYAK +R +
Sbjct: 183 IITNSDVTSIINSKEIQSILRPAGEKHEKRPFTQKKNPLRNNGVKIRLNPYAKILQRAEI 362
Query: 349 LAEAERVKSKKEKFDKKRKIVSKEEASA 376
+A +R K K KKR+ SA
Sbjct: 363 IAAEKRKAGKIIK--KKRQFKESTRTSA 440
>BQ151325
Length = 784
Score = 61.2 bits (147), Expect(2) = 1e-18
Identities = 35/75 (46%), Positives = 45/75 (59%), Gaps = 2/75 (2%)
Frame = +3
Query: 63 RKAGHQTSAESWGTGRA-VSRIPRVAGGGTHRAGQAAFGNMCRGGRMFAPTRIWRK-WHR 120
RKAG+QTS+ SWG SRI VAG GTHRA QAAF N GGRM+ T WHR
Sbjct: 246 RKAGNQTSSYSWGYRDVHESRITPVAGRGTHRAEQAAFSNKFHGGRMYDATPDTGS*WHR 425
Query: 121 KINVNQKRYAVVSAI 135
K ++ +++ ++
Sbjct: 426 KTHIGKQQNGTADSL 470
Score = 49.3 bits (116), Expect(2) = 1e-18
Identities = 21/35 (60%), Positives = 27/35 (77%)
Frame = +2
Query: 17 LESEMSTDSPNTLPIPDVMRASIRPDIVNFVHSNI 51
+E +M TD+ N LPI DVM A +RPDI+ F+HSNI
Sbjct: 104 VERDMDTDTTNCLPISDVMHAHLRPDIIYFIHSNI 208
>TC87023 similar to SP|O80361|RK4_TOBAC 50S ribosomal protein L4
chloroplast precursor (R-protein L4). [Common tobacco]
{Nicotiana tabacum}, partial (78%)
Length = 1283
Score = 47.4 bits (111), Expect = 1e-05
Identities = 45/132 (34%), Positives = 61/132 (46%), Gaps = 10/132 (7%)
Frame = +2
Query: 61 VSRKAGHQTSAESWGTGRAVSRIPRVAGGG--------THRAGQAAFGNMCR--GGRMFA 110
V R H + GT ++R V GGG T RA Q + R GG +F
Sbjct: 353 VHRAIIHDLQNKRRGTASTLTRA-EVRGGGRKPYNQKKTGRARQGSIRTPLRPGGGVIFG 529
Query: 111 PTRIWRKWHRKINVNQKRYAVVSAIAASAIPSLVLARGHRIETVPEFPLVVGDAAEGVEK 170
P R W KIN +KR A+ +A+A++A+ ++V V EF GD EG K
Sbjct: 530 PKP--RDWTIKINKKEKRLAISTAVASAAVNTVV---------VEEF----GDEFEGNAK 664
Query: 171 TKEAIKVLKQVG 182
TKE I +K+ G
Sbjct: 665 TKEFIAAMKRWG 700
>BG583329 similar to PIR|T00956|T00 translation initiation factor eIF-2 gamma
chain F20D22.6 - Arabidopsis thaliana, partial (47%)
Length = 680
Score = 30.0 bits (66), Expect = 1.7
Identities = 22/67 (32%), Positives = 33/67 (48%), Gaps = 1/67 (1%)
Frame = +2
Query: 252 GHLGRFVIWTKSAFEKLDSIYGTFDKGSEKKKGYVLPRAKMVNSDLTRI-INSDEVQSVV 310
GH RF + + F D +G KGSE K+G V AK+ S++ + I S + V
Sbjct: 479 GHCPRFSLSLRLTFSCFDGFFGVRTKGSE-KQGKV---AKLTKSEILMLTIGSMSTGAKV 646
Query: 311 RPIKKDV 317
+K D+
Sbjct: 647 TAVKNDL 667
>TC78462 similar to GP|21592780|gb|AAM64729.1 nucleic acid binding
protein-like {Arabidopsis thaliana}, partial (85%)
Length = 1281
Score = 28.9 bits (63), Expect = 3.8
Identities = 11/26 (42%), Positives = 17/26 (65%)
Frame = +1
Query: 115 WRKWHRKINVNQKRYAVVSAIAASAI 140
WR W RK+ +N +R VV A++ A+
Sbjct: 760 WRNWTRKMKMNMERPYVVHAVSIMAL 837
>AJ502494 similar to GP|7547109|gb| unknown protein {Arabidopsis thaliana},
partial (48%)
Length = 696
Score = 27.7 bits (60), Expect = 8.4
Identities = 16/48 (33%), Positives = 27/48 (55%), Gaps = 3/48 (6%)
Frame = +3
Query: 353 ERVKSKKEKFDKKRKIVSKEEASAIKAAGKAWYNTMVS---DSDYAEF 397
E VK+ D + K +K++A KAAG+AW + +++ + DY F
Sbjct: 393 ENVKAAIASSDVEHKAETKKKAVPRKAAGQAWEDPILAEWPEDDYRLF 536
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.131 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,705,704
Number of Sequences: 36976
Number of extensions: 109808
Number of successful extensions: 602
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 599
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 601
length of query: 408
length of database: 9,014,727
effective HSP length: 98
effective length of query: 310
effective length of database: 5,391,079
effective search space: 1671234490
effective search space used: 1671234490
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC123596.12


