

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123593.9 + phase: 0 /pseudo
(409 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
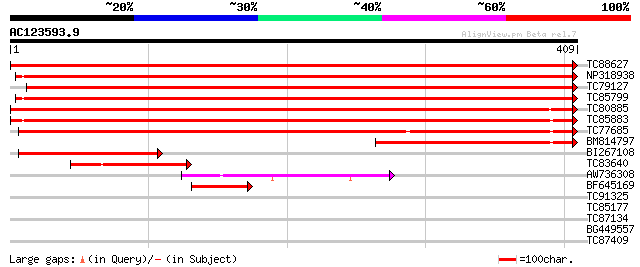
Score E
Sequences producing significant alignments: (bits) Value
TC88627 apyrase-like protein [Medicago truncatula] 817 0.0
NP318938 NP318938|AF288132.1|AAK15160.1 putative apyrase [Medica... 753 0.0
TC79127 apyrase-like protein [Medicago truncatula] 690 0.0
TC85799 apyrase-like protein [Medicago truncatula] 686 0.0
TC80885 apyrase-like protein [Medicago truncatula] 646 0.0
TC85883 apyrase-like protein [Medicago truncatula] 588 e-168
TC77685 apyrase-like protein [Medicago truncatula] 482 e-136
BM814797 similar to GP|10717159|gb apyrase 2 {Pisum sativum}, pa... 168 4e-42
BI267108 homologue to GP|10717159|gb apyrase 2 {Pisum sativum}, ... 114 5e-26
TC83640 similar to GP|14423550|gb|AAK62457.1 putative nucleoside... 70 2e-12
AW736308 similar to GP|19920124|gb| Putative nucleoside phosphat... 56 2e-08
BF645169 similar to GP|10717159|gb| apyrase 2 {Pisum sativum}, p... 53 2e-07
TC91325 similar to GP|19920124|gb|AAM08556.1 Putative nucleoside... 36 0.031
TC85177 similar to PIR|S56655|S56655 lipoxygenase (EC 1.13.11.12... 32 0.58
TC87134 similar to PIR|T48275|T48275 hypothetical protein T22P11... 30 1.7
BG449557 similar to GP|19920124|gb| Putative nucleoside phosphat... 29 2.9
TC87409 similar to PIR|F96686|F96686 unknown protein F15E12.7 [i... 28 4.9
>TC88627 apyrase-like protein [Medicago truncatula]
Length = 1614
Score = 817 bits (2111), Expect = 0.0
Identities = 405/409 (99%), Positives = 406/409 (99%)
Frame = +1
Query: 1 MEFLITLITTVLLLLMPAITSSQYLGNNLLTNRKIFQKQETISSYAVVFDAGSTGSRIHV 60
MEFLITLITTVLLLLMPAITSSQYLGNNLLTNRKIFQKQETISSYAVVFDAGSTGSRIHV
Sbjct: 73 MEFLITLITTVLLLLMPAITSSQYLGNNLLTNRKIFQKQETISSYAVVFDAGSTGSRIHV 252
Query: 61 YHFDQNLDLLHIGKDVEFFNKITPGLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHKTP 120
YHFDQNLDLLHIGKDVEFFNKITPGLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHKTP
Sbjct: 253 YHFDQNLDLLHIGKDVEFFNKITPGLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHKTP 432
Query: 121 IRLGATAGLRLLNGDASEKILQAVRDMFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYA 180
IRLGATAGLRLLNGDASEKILQAVRDMFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYA
Sbjct: 433 IRLGATAGLRLLNGDASEKILQAVRDMFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYA 612
Query: 181 LGNLGKKYTKTVGVMDLGGGSVQMAYAVSKKTAKNAPKVADGVDPYIKKLVLKGKPYDLY 240
LGNLGKKYTKTVGVMDLGGGSVQMAYAVSKKTAKNAPKVADGVDPYIKKLVLKGKPYDLY
Sbjct: 613 LGNLGKKYTKTVGVMDLGGGSVQMAYAVSKKTAKNAPKVADGVDPYIKKLVLKGKPYDLY 792
Query: 241 VHSYLHFGREASRAEIMKVTRSSPNPCLLAGFDGTYTYAGEEFKAKAPASGANFNGCKKI 300
VHSYLHFGREASRAEIMKVTRSSPN C+L GFDGTYTYAGEEFKAKAPASGANFNGCKKI
Sbjct: 793 VHSYLHFGREASRAEIMKVTRSSPNTCILTGFDGTYTYAGEEFKAKAPASGANFNGCKKI 972
Query: 301 IRKALKLNYPCPYQNCTFGGIWNGGGGNGQKHLFASSSFFYLPEDVGMVDPKTPNFKIRP 360
IRKALKLNYPCPYQNCTFGGIWNGGGGNGQKHL ASSSFFYLPEDVGMVDPKTPNFKIRP
Sbjct: 973 IRKALKLNYPCPYQNCTFGGIWNGGGGNGQKHLLASSSFFYLPEDVGMVDPKTPNFKIRP 1152
Query: 361 VDLVSEAKKACALNFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDGF 409
VDLVSEAKKACALNFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDGF
Sbjct: 1153VDLVSEAKKACALNFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDGF 1299
>NP318938 NP318938|AF288132.1|AAK15160.1 putative apyrase [Medicago
truncatula]
Length = 1401
Score = 753 bits (1943), Expect = 0.0
Identities = 373/406 (91%), Positives = 391/406 (95%), Gaps = 1/406 (0%)
Frame = +1
Query: 5 ITLITTVLLLLMPAITSSQYLGNN-LLTNRKIFQKQETISSYAVVFDAGSTGSRIHVYHF 63
+TLIT LL +MP+I+ SQYLGNN LLTNRKIF KQE ISSYAVVFDAGSTGSR+HVYHF
Sbjct: 43 MTLIT-FLLFIMPSISYSQYLGNNILLTNRKIFPKQEPISSYAVVFDAGSTGSRVHVYHF 219
Query: 64 DQNLDLLHIGKDVEFFNKITPGLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHKTPIRL 123
DQNL+LLH+GKDVEF+NK TPGLS+YA++PE+AAKSLIPLL+QAE+VVP D KTPIRL
Sbjct: 220 DQNLNLLHVGKDVEFYNKTTPGLSAYADNPEEAAKSLIPLLEQAESVVPEDQRSKTPIRL 399
Query: 124 GATAGLRLLNGDASEKILQAVRDMFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYALGN 183
GATAGLRLLNGDASEKILQ+VRD+FSNRSTFNVQPDAVSIIDGTQEG YLWVTVNYALGN
Sbjct: 400 GATAGLRLLNGDASEKILQSVRDLFSNRSTFNVQPDAVSIIDGTQEGCYLWVTVNYALGN 579
Query: 184 LGKKYTKTVGVMDLGGGSVQMAYAVSKKTAKNAPKVADGVDPYIKKLVLKGKPYDLYVHS 243
LGKK+TKTVGVMD+GGGSVQMAYAVSK TAKNAPKVADG DPYIKKLVLKGKPYDLYVHS
Sbjct: 580 LGKKFTKTVGVMDVGGGSVQMAYAVSKYTAKNAPKVADGEDPYIKKLVLKGKPYDLYVHS 759
Query: 244 YLHFGREASRAEIMKVTRSSPNPCLLAGFDGTYTYAGEEFKAKAPASGANFNGCKKIIRK 303
YLHFGREASRAEIMKVTRSSPNPCLLAGFDGTYTYAGEEFKAKAPASGANFNGCKKIIRK
Sbjct: 760 YLHFGREASRAEIMKVTRSSPNPCLLAGFDGTYTYAGEEFKAKAPASGANFNGCKKIIRK 939
Query: 304 ALKLNYPCPYQNCTFGGIWNGGGGNGQKHLFASSSFFYLPEDVGMVDPKTPNFKIRPVDL 363
ALKLNYPCPYQNCTFGGIWNGGGGNGQKHLFASSSFFYLPEDVGMVDPKTPNFKIRPVDL
Sbjct: 940 ALKLNYPCPYQNCTFGGIWNGGGGNGQKHLFASSSFFYLPEDVGMVDPKTPNFKIRPVDL 1119
Query: 364 VSEAKKACALNFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDGF 409
VSEAKKACALNFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDGF
Sbjct: 1120VSEAKKACALNFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDGF 1257
>TC79127 apyrase-like protein [Medicago truncatula]
Length = 1501
Score = 690 bits (1780), Expect = 0.0
Identities = 339/398 (85%), Positives = 367/398 (92%), Gaps = 1/398 (0%)
Frame = +3
Query: 13 LLLMPAITSSQYLGNN-LLTNRKIFQKQETISSYAVVFDAGSTGSRIHVYHFDQNLDLLH 71
L +MP+I+ SQ LGNN LLTNRKIF KQETISSYAVVFDAGSTGSR+HVYHFDQNL+LLH
Sbjct: 87 LFIMPSISYSQNLGNNILLTNRKIFPKQETISSYAVVFDAGSTGSRVHVYHFDQNLNLLH 266
Query: 72 IGKDVEFFNKITPGLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHKTPIRLGATAGLRL 131
+GKDVEF+NK TPGLS+YA++PEQAAKSLIPLL+QAE+VVP D KTP+RLGATAGLRL
Sbjct: 267 VGKDVEFYNKTTPGLSAYADNPEQAAKSLIPLLEQAESVVPEDQRSKTPVRLGATAGLRL 446
Query: 132 LNGDASEKILQAVRDMFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYALGNLGKKYTKT 191
LNGDASEKILQ+VRD+ SNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYALG LGKK+TKT
Sbjct: 447 LNGDASEKILQSVRDLLSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYALGTLGKKFTKT 626
Query: 192 VGVMDLGGGSVQMAYAVSKKTAKNAPKVADGVDPYIKKLVLKGKPYDLYVHSYLHFGREA 251
VGVMDLGGGSVQMAYAVS+ TAKNAPKVADG DPYIKKLVLKGK YDLYVHSYLHFG EA
Sbjct: 627 VGVMDLGGGSVQMAYAVSRNTAKNAPKVADGDDPYIKKLVLKGKKYDLYVHSYLHFGTEA 806
Query: 252 SRAEIMKVTRSSPNPCLLAGFDGTYTYAGEEFKAKAPASGANFNGCKKIIRKALKLNYPC 311
SRAEI+KVT +SPNPC+LAGFDGTY YAGEEFKA A ASGA+F CKKI+ +ALKLNYPC
Sbjct: 807 SRAEILKVTHNSPNPCILAGFDGTYRYAGEEFKANALASGASFKKCKKIVHQALKLNYPC 986
Query: 312 PYQNCTFGGIWNGGGGNGQKHLFASSSFFYLPEDVGMVDPKTPNFKIRPVDLVSEAKKAC 371
PYQNCTFGGIWNGGGG+GQ+ LFA+S FFYL +VGMVDP PNFKIRPVD SEAKKAC
Sbjct: 987 PYQNCTFGGIWNGGGGSGQRKLFAASFFFYLAAEVGMVDPNKPNFKIRPVDFESEAKKAC 1166
Query: 372 ALNFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDGF 409
ALNFEDAKS+YPFLAKKNIASYVCMDLIYQYVLLVDGF
Sbjct: 1167ALNFEDAKSSYPFLAKKNIASYVCMDLIYQYVLLVDGF 1280
>TC85799 apyrase-like protein [Medicago truncatula]
Length = 1674
Score = 686 bits (1770), Expect = 0.0
Identities = 340/407 (83%), Positives = 372/407 (90%), Gaps = 2/407 (0%)
Frame = +3
Query: 5 ITLITTVLLLLMPAITSSQYLGNN-LLTNRKIFQKQETISSYAVVFDAGSTGSRIHVYHF 63
+TLIT LL +MP+I+ SQYLGNN LLTNRKIF KQE ISSYAVVFDAGSTGSR+HVYHF
Sbjct: 216 MTLIT-FLLFIMPSISYSQYLGNNILLTNRKIFPKQEPISSYAVVFDAGSTGSRVHVYHF 392
Query: 64 DQNLDLLHIGKDVEFFNKITPGLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHKTPIRL 123
DQNL+LLH+GKDVEF+NK TPGLS+YA++PE+AAKSLIPLL+QAE+VVP D KTPIRL
Sbjct: 393 DQNLNLLHVGKDVEFYNKTTPGLSAYADNPEEAAKSLIPLLEQAESVVPEDQRSKTPIRL 572
Query: 124 GATAGLRLLNGDASEKILQAVRDMFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYALGN 183
GATAGLRLLNGDASEKILQ+VRD+FSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYALG
Sbjct: 573 GATAGLRLLNGDASEKILQSVRDLFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYALGK 752
Query: 184 LGKKYTKTVGVMDLGGGSVQMAYAVSKKTAKNAPKVADGVDPYIKKLVLKGKPYDLYVHS 243
LGKK+TKTVGVMDLGGGSVQMAYAVSK TAKNAPKVADG DPYIKKLVLKGK YDLYVHS
Sbjct: 753 LGKKFTKTVGVMDLGGGSVQMAYAVSKYTAKNAPKVADGEDPYIKKLVLKGKKYDLYVHS 932
Query: 244 YLHFGREASRAEIMKVTRSSPNPCLLAGFDGTYTYAGEEFKAKAPASGANFNGCKKIIRK 303
YLHFGREASRAEI+KVT +SPNPC+LAGFDGTYTYAGEEFKA APASGANF CKKI+R+
Sbjct: 933 YLHFGREASRAEILKVTHNSPNPCILAGFDGTYTYAGEEFKANAPASGANFKKCKKIVRE 1112
Query: 304 ALKLNYPCPYQNCTFGGIWNGGGGNGQKHLFASSSFFYLPEDVGMVDPKTP-NFKIRPVD 362
ALKLNYPCPYQNCTFGGIW+GGGG+GQ+ LFA+SSFFYL ED+G+VDP TP + +RPVD
Sbjct: 1113ALKLNYPCPYQNCTFGGIWSGGGGSGQRILFAASSFFYLAEDIGLVDPNTPYSLTLRPVD 1292
Query: 363 LVSEAKKACALNFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDGF 409
L +EAKKAC LN E+AKSTYP L NI YVCMDLIYQYVLLVDGF
Sbjct: 1293LETEAKKACTLNLEEAKSTYPLLVDFNIVEYVCMDLIYQYVLLVDGF 1433
>TC80885 apyrase-like protein [Medicago truncatula]
Length = 1451
Score = 646 bits (1666), Expect = 0.0
Identities = 311/409 (76%), Positives = 359/409 (87%)
Frame = +2
Query: 1 MEFLITLITTVLLLLMPAITSSQYLGNNLLTNRKIFQKQETISSYAVVFDAGSTGSRIHV 60
MEFLITLITT LLLLMPAITSSQYLGNNLLTNRKIFQKQET++SYAV+FDAGSTG+R+HV
Sbjct: 5 MEFLITLITTFLLLLMPAITSSQYLGNNLLTNRKIFQKQETLTSYAVIFDAGSTGTRVHV 184
Query: 61 YHFDQNLDLLHIGKDVEFFNKITPGLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHKTP 120
YHFDQNLDLLHIG D+EF +KI PGLS+YA++PEQAAKSL+PLL++AE+V+P D+H KTP
Sbjct: 185 YHFDQNLDLLHIGNDIEFVDKIKPGLSAYADNPEQAAKSLLPLLEEAEDVIPEDMHPKTP 364
Query: 121 IRLGATAGLRLLNGDASEKILQAVRDMFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYA 180
+RLGATAGLRLLNGDA+EKILQA R+MFSNRST NVQ DAVSIIDGTQEGSY+WVTVNY
Sbjct: 365 LRLGATAGLRLLNGDAAEKILQATRNMFSNRSTLNVQSDAVSIIDGTQEGSYMWVTVNYI 544
Query: 181 LGNLGKKYTKTVGVMDLGGGSVQMAYAVSKKTAKNAPKVADGVDPYIKKLVLKGKPYDLY 240
LGNLGK +TKTVGV+DLGGGSVQM YAVSKKTAKNAPKVADG DPYIKKLVLKGK YDLY
Sbjct: 545 LGNLGKSFTKTVGVIDLGGGSVQMTYAVSKKTAKNAPKVADGEDPYIKKLVLKGKQYDLY 724
Query: 241 VHSYLHFGREASRAEIMKVTRSSPNPCLLAGFDGTYTYAGEEFKAKAPASGANFNGCKKI 300
VHSYL FG+EA+RA+++ T S NPC+L GF+GT+TY+G E+KA +P+SG+NFN CK+I
Sbjct: 725 VHSYLRFGKEATRAQVLNATNGSANPCILPGFNGTFTYSGVEYKAFSPSSGSNFNECKEI 904
Query: 301 IRKALKLNYPCPYQNCTFGGIWNGGGGNGQKHLFASSSFFYLPEDVGMVDPKTPNFKIRP 360
I K LK+N PCPY +CTF GIWNGGGG+GQK LF +S+F YL EDVGMV+P PN + P
Sbjct: 905 ILKVLKVNDPCPYSSCTFSGIWNGGGGSGQKKLFVTSAFAYLTEDVGMVEPNKPNSTLHP 1084
Query: 361 VDLVSEAKKACALNFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDGF 409
+D EAK+ACALNFED KSTYP L + YVCMDL+YQ+VLLV GF
Sbjct: 1085IDFEIEAKRACALNFEDVKSTYPRLTEAK-RPYVCMDLLYQHVLLVHGF 1228
>TC85883 apyrase-like protein [Medicago truncatula]
Length = 1848
Score = 588 bits (1516), Expect = e-168
Identities = 284/409 (69%), Positives = 343/409 (83%)
Frame = +3
Query: 1 MEFLITLITTVLLLLMPAITSSQYLGNNLLTNRKIFQKQETISSYAVVFDAGSTGSRIHV 60
MEFLI LIT LL LMP I+SSQYLGNN+LTNRKIF KQET++SYAVVFDAGSTGSR+HV
Sbjct: 246 MEFLIKLIT-FLLFLMPTISSSQYLGNNILTNRKIFPKQETLTSYAVVFDAGSTGSRVHV 422
Query: 61 YHFDQNLDLLHIGKDVEFFNKITPGLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHKTP 120
YHFDQNLDLLHIG DVEF+NK TPGLS+YA++P++AA+SLIPLL+QAE VVP++L KTP
Sbjct: 423 YHFDQNLDLLHIGNDVEFYNKTTPGLSAYADNPKEAAESLIPLLEQAERVVPVNLQPKTP 602
Query: 121 IRLGATAGLRLLNGDASEKILQAVRDMFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYA 180
++LGATAGLRLL+G++SE IL+AV + RSTFNVQ DAV IIDGTQEGSYLWVT+NY
Sbjct: 603 VKLGATAGLRLLDGNSSELILEAVSSLLKKRSTFNVQSDAVGIIDGTQEGSYLWVTINYV 782
Query: 181 LGNLGKKYTKTVGVMDLGGGSVQMAYAVSKKTAKNAPKVADGVDPYIKKLVLKGKPYDLY 240
LGNLGK +++TV V DLGGGSVQM YAVS++ AK AP+V G DPYIKK+VLKGK Y LY
Sbjct: 783 LGNLGKDFSETVAVADLGGGSVQMVYAVSREQAKKAPQVPQGEDPYIKKIVLKGKKYYLY 962
Query: 241 VHSYLHFGREASRAEIMKVTRSSPNPCLLAGFDGTYTYAGEEFKAKAPASGANFNGCKKI 300
VHSYL FG+EASRAEI+KVT SPNPC+LAG+ GTYTY+GEE+KA +PASG+NF+ CK+I
Sbjct: 963 VHSYLRFGKEASRAEILKVTNGSPNPCILAGYHGTYTYSGEEYKAFSPASGSNFDECKEI 1142
Query: 301 IRKALKLNYPCPYQNCTFGGIWNGGGGNGQKHLFASSSFFYLPEDVGMVDPKTPNFKIRP 360
I KALK+N PCPY C+FGGIWNGGGG+GQK L+ +SSF+Y+P V + DP PN KIR
Sbjct: 1143ILKALKVNDPCPYGKCSFGGIWNGGGGSGQKTLYVTSSFYYVPTGVNIADPNKPNSKIRI 1322
Query: 361 VDLVSEAKKACALNFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDGF 409
DL + A++ C ++DAK+TYP + + ++ Y C+DLIYQY L VDGF
Sbjct: 1323EDLKTGAEQVCKTKYKDAKATYPLIYEDSL-PYACLDLIYQYTLFVDGF 1466
>TC77685 apyrase-like protein [Medicago truncatula]
Length = 2172
Score = 482 bits (1241), Expect = e-136
Identities = 233/404 (57%), Positives = 306/404 (75%), Gaps = 1/404 (0%)
Frame = +1
Query: 7 LITTVLLLLMPAITSSQYLGNNLLTNRKIFQKQETISSYAVVFDAGSTGSRIHVYHFDQN 66
L+ T +L +MP+ ++ G+ L NRK+ +++ SYAV+FDAGS+GSR+HV+HFDQN
Sbjct: 433 LLITFVLYMMPSSSNYDSAGDYALLNRKLSPDKKSGGSYAVIFDAGSSGSRVHVFHFDQN 612
Query: 67 LDLLHIGKDVEFFNKITPGLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHKTPIRLGAT 126
LDL+HIGKD+E F ++ PGLS+YA P+QAA+SL+ LL++AE VVP +L KTP+R+GAT
Sbjct: 613 LDLVHIGKDLELFEQLKPGLSAYAQKPQQAAESLVSLLEKAEGVVPRELRSKTPVRIGAT 792
Query: 127 AGLRLLNGDASEKILQAVRDMFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYALGNLGK 186
AGLR L GDAS+KIL+AVRD+ +RS+F DAV+++DGTQEG+Y WVT+NY LGNLGK
Sbjct: 793 AGLRALEGDASDKILRAVRDLLKHRSSFKSDADAVTVLDGTQEGAYQWVTINYLLGNLGK 972
Query: 187 KYTKTVGVMDLGGGSVQMAYAVSKKTAKNAPKVADGVDPYIKKLVLKGKPYDLYVHSYLH 246
Y+KTVGV+DLGGGSVQMAYA+S+ A AP+V DG DPY+K++ L+G+ Y LYVHSYL
Sbjct: 973 DYSKTVGVVDLGGGSVQMAYAISESEAAMAPQVMDGEDPYVKEMFLRGRKYYLYVHSYLR 1152
Query: 247 FGREASRAEIMKVTRSSPNPCLLAGFDGTYTYAGEEFKAKAPASGANFNGCKKIIRKALK 306
+G A+RAEI+KV + NPC+L+G DGTY Y G+ FKA +SGA+ N CK + KALK
Sbjct: 1153YGLLAARAEILKVAGDAENPCILSGSDGTYKYGGKSFKAS--SSGASLNECKSVAHKALK 1326
Query: 307 LN-YPCPYQNCTFGGIWNGGGGNGQKHLFASSSFFYLPEDVGMVDPKTPNFKIRPVDLVS 365
+N C + CTFGGIWNGGGG+GQK+LF +S FF + G VDP +P +RP D
Sbjct: 1327VNESTCTHMKCTFGGIWNGGGGDGQKNLFVASFFFDRAAEAGFVDPNSPVAIVRPADFED 1506
Query: 366 EAKKACALNFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDGF 409
AK+AC E+AKSTYP + + N+ Y+CMDL+YQY LLVDGF
Sbjct: 1507AAKQACQTKLENAKSTYPRVEEGNL-PYLCMDLVYQYTLLVDGF 1635
>BM814797 similar to GP|10717159|gb apyrase 2 {Pisum sativum}, partial (42%)
Length = 690
Score = 168 bits (425), Expect = 4e-42
Identities = 79/146 (54%), Positives = 100/146 (68%), Gaps = 1/146 (0%)
Frame = +1
Query: 265 NPCLLAGFDGTYTYAGEEFKAKAPASGANFNGCKKIIRKALKLNYP-CPYQNCTFGGIWN 323
NPC+LAG+DG+Y Y G+ F A + SG++ N CK I AL +N C + CTFGGIWN
Sbjct: 25 NPCILAGYDGSYKYGGKSFNASSSPSGSSLNECKSIALNALTVNESTCAHMKCTFGGIWN 204
Query: 324 GGGGNGQKHLFASSSFFYLPEDVGMVDPKTPNFKIRPVDLVSEAKKACALNFEDAKSTYP 383
GGGG+GQK+LF +S FF G +PK+P K+RPVD + AK+AC EDAKSTYP
Sbjct: 205 GGGGDGQKNLFVASFFFDRAAQAGFANPKSPVAKVRPVDFKNAAKQACQTKLEDAKSTYP 384
Query: 384 FLAKKNIASYVCMDLIYQYVLLVDGF 409
+ N+ Y+CMDL+YQY LLVDGF
Sbjct: 385 LVDDGNL-PYLCMDLVYQYTLLVDGF 459
>BI267108 homologue to GP|10717159|gb apyrase 2 {Pisum sativum}, partial
(24%)
Length = 449
Score = 114 bits (286), Expect = 5e-26
Identities = 55/104 (52%), Positives = 81/104 (77%)
Frame = +2
Query: 7 LITTVLLLLMPAITSSQYLGNNLLTNRKIFQKQETISSYAVVFDAGSTGSRIHVYHFDQN 66
L T++L +MP+ +S++ + + LT+RKI ++ S+AVVFDAGS+GSR+HV+ FD+N
Sbjct: 122 LXVTLILYIMPSTSSNESIEDYALTHRKISPDRKISDSFAVVFDAGSSGSRVHVFRFDRN 301
Query: 67 LDLLHIGKDVEFFNKITPGLSSYANDPEQAAKSLIPLLQQAENV 110
L+L+ IG D+E F +I PGLS+YA DP+QAAKSL+ LL +AE+V
Sbjct: 302 LELVKIGNDLEVFLQIKPGLSAYARDPQQAAKSLVSLLDKAESV 433
>TC83640 similar to GP|14423550|gb|AAK62457.1 putative nucleoside
triphosphatase {Arabidopsis thaliana}, partial (12%)
Length = 601
Score = 69.7 bits (169), Expect = 2e-12
Identities = 34/87 (39%), Positives = 55/87 (63%)
Frame = +3
Query: 45 YAVVFDAGSTGSRIHVYHFDQNLDLLHIGKDVEFFNKITPGLSSYANDPEQAAKSLIPLL 104
Y ++ D GSTG+R+HV+ + + L GK ++ PGLSS+ NDP+ A +SL+ ++
Sbjct: 342 YRIIIDGGSTGTRVHVFKYKMK-NALDFGKKGLVSMRVNPGLSSFGNDPDGAGRSLLEVV 518
Query: 105 QQAENVVPIDLHHKTPIRLGATAGLRL 131
A+ +P + +T IRL ATAG+R+
Sbjct: 519 DFAKRRIPKENWRETEIRLMATAGMRM 599
>AW736308 similar to GP|19920124|gb| Putative nucleoside phosphatase {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (3%)
Length = 552
Score = 56.2 bits (134), Expect = 2e-08
Identities = 39/159 (24%), Positives = 72/159 (44%), Gaps = 6/159 (3%)
Frame = +1
Query: 125 ATAGLRLLNGDASEKILQAVRDMFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYALGNL 184
ATAG+R ++ D ++L+ V + + S F + ++ G +E Y WV +NY +G
Sbjct: 7 ATAGMRRIHRDDVFRVLEDVEAVAKDHS-FMFDKRWIRVLSGREEAYYGWVALNYNMGRF 183
Query: 185 GKKY---TKTVGVMDLGGGSVQMAYAVSKKTAKNAPKVADGVDPYIKKLVLKGKPYDLYV 241
Y + T+G++DLGG S+Q+ + + T + + ++V P
Sbjct: 184 DDDYHRGSSTLGLVDLGGSSLQIVVEIDRDTGDDVNAIRSEFGSIEHRIVAYSLPSFGLN 363
Query: 242 HSY---LHFGREASRAEIMKVTRSSPNPCLLAGFDGTYT 277
++ + R R E + +PCL++ F YT
Sbjct: 364 EAFDRTVAMLRNNQRVESTRGVSELRHPCLMSTFVQNYT 480
>BF645169 similar to GP|10717159|gb| apyrase 2 {Pisum sativum}, partial (10%)
Length = 542
Score = 52.8 bits (125), Expect = 2e-07
Identities = 23/44 (52%), Positives = 31/44 (70%)
Frame = +1
Query: 132 LNGDASEKILQAVRDMFSNRSTFNVQPDAVSIIDGTQEGSYLWV 175
L GD S++ILQAV D+ RST +P+ V + DGTQEG++ WV
Sbjct: 235 LEGDVSDRILQAV*DLLKQRSTLKSEPNVVDVQDGTQEGAFQWV 366
>TC91325 similar to GP|19920124|gb|AAM08556.1 Putative nucleoside
phosphatase {Oryza sativa} [Oryza sativa (japonica
cultivar-group)], partial (2%)
Length = 672
Score = 35.8 bits (81), Expect = 0.031
Identities = 37/125 (29%), Positives = 53/125 (41%), Gaps = 23/125 (18%)
Frame = +2
Query: 3 FLITLITTVLLLLMPAITSSQYLGNNLLTNRKIFQKQETISSYAVVFDAGSTGSRIHVYH 62
F IT + +L+LL +LG L ++ K + S Y VV D GSTG+R++VY
Sbjct: 323 FKITAVIFILILL--------FLGFYLESHN---WKPNSTSYYTVVIDCGSTGTRVNVYE 469
Query: 63 F-------DQNLD-LLHIGKDVEFFNK---------------ITPGLSSYANDPEQAAKS 99
+ +NL LLH D K PGL ND + ++
Sbjct: 470 WMLGGVVGKKNLPILLHSYPDNNATKKNNSLWKNSCQYHCMQTEPGLDKLVNDSLRVRRA 649
Query: 100 LIPLL 104
L PL+
Sbjct: 650 LEPLI 664
>TC85177 similar to PIR|S56655|S56655 lipoxygenase (EC 1.13.11.12) loxG -
garden pea, partial (75%)
Length = 2646
Score = 31.6 bits (70), Expect = 0.58
Identities = 18/66 (27%), Positives = 33/66 (49%), Gaps = 9/66 (13%)
Frame = +3
Query: 310 PCPYQNCTFGGI---WNGGGGNGQKHLF------ASSSFFYLPEDVGMVDPKTPNFKIRP 360
PC Y+ G + G G+KHL + S++FY+P D ++ PK+ +F +
Sbjct: 645 PCLYRPVLGGSTALPYPRRGRTGRKHLEKYPETESRSNYFYIPRDELIIPPKSSDFVVNT 824
Query: 361 VDLVSE 366
+ +S+
Sbjct: 825 IKSISQ 842
>TC87134 similar to PIR|T48275|T48275 hypothetical protein T22P11.130 -
Arabidopsis thaliana, partial (87%)
Length = 1374
Score = 30.0 bits (66), Expect = 1.7
Identities = 17/71 (23%), Positives = 36/71 (49%)
Frame = +3
Query: 21 SSQYLGNNLLTNRKIFQKQETISSYAVVFDAGSTGSRIHVYHFDQNLDLLHIGKDVEFFN 80
++ +LG+ LLTN + + ++T + + + S H Y +++ + L +I + + +
Sbjct: 651 ATNHLGHFLLTNLLLEKMKQTAKATGIEGRIINLSSIAHTYTYEEGIRLDNINDQIGYSD 830
Query: 81 KITPGLSSYAN 91
K G S AN
Sbjct: 831 KKAYGQSKLAN 863
>BG449557 similar to GP|19920124|gb| Putative nucleoside phosphatase {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (4%)
Length = 662
Score = 29.3 bits (64), Expect = 2.9
Identities = 18/60 (30%), Positives = 30/60 (50%)
Frame = +1
Query: 3 FLITLITTVLLLLMPAITSSQYLGNNLLTNRKIFQKQETISSYAVVFDAGSTGSRIHVYH 62
F + ++ T+ LLL + Y N+ + + Y VV D GSTG+R++VY+
Sbjct: 376 FRLVILLTLFLLLSYIVFMFVYSFWNIGSGK-----------YYVVLDCGSTGTRVYVYN 522
>TC87409 similar to PIR|F96686|F96686 unknown protein F15E12.7 [imported] -
Arabidopsis thaliana, partial (84%)
Length = 2112
Score = 28.5 bits (62), Expect = 4.9
Identities = 17/47 (36%), Positives = 25/47 (53%)
Frame = +2
Query: 336 SSSFFYLPEDVGMVDPKTPNFKIRPVDLVSEAKKACALNFEDAKSTY 382
SSSFF LP + + P+ P+F + P D +A C ++ A TY
Sbjct: 500 SSSFFVLPCNHPLCKPRVPDFSL-PTD--CDANSLCHYSYFYADGTY 631
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.137 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,158,394
Number of Sequences: 36976
Number of extensions: 168872
Number of successful extensions: 932
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 904
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 919
length of query: 409
length of database: 9,014,727
effective HSP length: 98
effective length of query: 311
effective length of database: 5,391,079
effective search space: 1676625569
effective search space used: 1676625569
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC123593.9


