

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123593.8 + phase: 0
(550 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
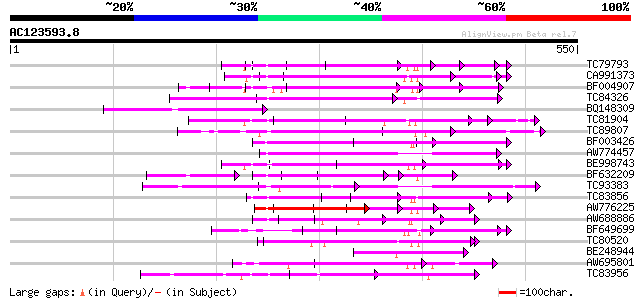
Score E
Sequences producing significant alignments: (bits) Value
TC79793 weakly similar to GP|6630464|gb|AAF19552.1| F23N19.4 {Ar... 123 2e-28
CA991373 weakly similar to PIR|D86260|D86 protein T12C24.22 [imp... 118 7e-27
BF004907 weakly similar to GP|8953393|emb putative protein {Arab... 111 8e-25
TC84326 weakly similar to PIR|F85225|F85225 hypothetical protein... 104 8e-23
BQ148309 weakly similar to GP|6721172|gb| hypothetical protein {... 92 4e-19
TC81904 weakly similar to GP|8778410|gb|AAF79418.1| F16A14.3 {Ar... 89 3e-18
TC89807 weakly similar to PIR|B96600|B96600 protein F14J16.14 [i... 89 6e-18
BF003426 similar to PIR|F96760|F96 hypothetical protein T9L24.39... 88 7e-18
AW774457 weakly similar to GP|10176973|dbj gb|AAF19552.1~gene_id... 88 1e-17
BE998743 similar to GP|22128591|gb| fertility restorer-like prot... 87 2e-17
BF632209 weakly similar to GP|8493579|gb|A Contains a RepB PF|01... 82 7e-16
TC93383 similar to PIR|T47786|T47786 hypothetical protein F17J16... 80 3e-15
TC83856 weakly similar to PIR|B96656|B96656 unknown protein 419... 78 1e-14
AW776225 weakly similar to GP|18461169|dbj hypothetical protein~... 77 2e-14
AW688886 similar to GP|3258568|gb|A Unknown protein {Arabidopsis... 76 4e-14
BF649699 similar to GP|9757911|dbj gb|AAF19552.1~gene_id:MMN10.1... 75 8e-14
TC80520 weakly similar to PIR|T02562|T02562 probable salt-induci... 74 1e-13
BE248944 similar to GP|11994509|dbj gb|AAF27119.1~gene_id:F4B12.... 72 4e-13
AW695801 weakly similar to PIR|T01622|T01 probable salt-inducibl... 70 2e-12
TC83956 weakly similar to GP|15982931|gb|AAL09812.1 AT3g53700/F4... 68 1e-11
>TC79793 weakly similar to GP|6630464|gb|AAF19552.1| F23N19.4 {Arabidopsis
thaliana}, partial (7%)
Length = 1580
Score = 123 bits (308), Expect = 2e-28
Identities = 73/241 (30%), Positives = 132/241 (54%), Gaps = 5/241 (2%)
Frame = +1
Query: 206 FQGLLSALCRYKNVQDAEHLL--FCNKNVFPLDTKSFNIILNGWCNLIVSARNAERIWEE 263
+ L+ C K V A+ + V P D +S++I++NG+C I A +++E
Sbjct: 565 YNSLMDGYCLVKEVNKAKSIFNTMAQGGVNP-DIQSYSIMINGFCK-IKKFDEAMNLFKE 738
Query: 264 MSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYNAVIFSLAKNRM 323
M ++ I DVV+Y+S+I SKS ++ LQL +QM R + P+ YN+++ +L K
Sbjct: 739 MHRKNIIPDVVTYSSLIDGLSKSGRISYALQLVDQMHDRGVPPNICTYNSILDALCKTHQ 918
Query: 324 VKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRTFHA 383
V +A+ L+ K +D PD TY+ LIK LC++ K+++A+++F +L +G + ++ T+
Sbjct: 919 VDKAIALLTKFKDKGFQPDISTYSILIKGLCQSGKLEDARKVFEGLLVKGHNLNVDTYTI 1098
Query: 384 F---FRILRVKEEVFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDG 440
F + + E LL KM++ GC P +TY ++I + + D +++ M G
Sbjct: 1099MIQGFCVEGLFNEALALLSKMEDNGCIPDAKTYEIIILSLFKKDENDMAEKLLREMIARG 1278
Query: 441 I 441
+
Sbjct: 1279L 1281
Score = 118 bits (295), Expect = 7e-27
Identities = 68/250 (27%), Positives = 133/250 (53%), Gaps = 3/250 (1%)
Frame = +1
Query: 229 NKNVFPLDTKSFNIILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSK 288
NK + P + ++N +++G+C L+ A+ I+ M++ + D+ SY+ +I+ + K K
Sbjct: 535 NKGIKP-NFVTYNSLMDGYC-LVKEVNKAKSIFNTMAQGGVNPDIQSYSIMINGFCKIKK 708
Query: 289 LYRVLQLFEQMKKRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNS 348
+ LF++M ++NI PD Y+++I L+K+ + A+ L+ +M D V P+ TYNS
Sbjct: 709 FDEAMNLFKEMHRKNIIPDVVTYSSLIDGLSKSGRISYALQLVDQMHDRGVPPNICTYNS 888
Query: 349 LIKPLCKARKIDEAKEIFNVMLERGISPSIRTFHAFFRILRVK---EEVFELLDKMKELG 405
++ LCK ++D+A + ++G P I T+ + L E+ ++ + + G
Sbjct: 889 ILDALCKTHQVDKAIALLTKFKDKGFQPDISTYSILIKGLCQSGKLEDARKVFEGLLVKG 1068
Query: 406 CNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAY 465
N ++TY ++I+ FC +E + + M ++G D +Y ++I LF + + A
Sbjct: 1069HNLNVDTYTIMIQGFCVEGLFNEALALLSKMEDNGCIPDAKTYEIIILSLFKKDENDMAE 1248
Query: 466 KYYIEMQEKG 475
K EM +G
Sbjct: 1249KLLREMIARG 1278
Score = 110 bits (275), Expect = 1e-24
Identities = 61/221 (27%), Positives = 117/221 (52%), Gaps = 3/221 (1%)
Frame = +1
Query: 269 IQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYNAVIFSLAKNRMVKEAV 328
I+ + V+Y S++ Y ++ + +F M + + PD + Y+ +I K + EA+
Sbjct: 544 IKPNFVTYNSLMDGYCLVKEVNKAKSIFNTMAQGGVNPDIQSYSIMINGFCKIKKFDEAM 723
Query: 329 NLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRTFHAFFRIL 388
NL +M N+ PD +TY+SLI L K+ +I A ++ + M +RG+ P+I T+++ L
Sbjct: 724 NLFKEMHRKNIIPDVVTYSSLIDGLSKSGRISYALQLVDQMHDRGVPPNICTYNSILDAL 903
Query: 389 RVKEEV---FELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDGIGHDR 445
+V LL K K+ G P I TY +LI+ C+ +L++ ++++ + G +
Sbjct: 904 CKTHQVDKAIALLTKFKDKGFQPDISTYSILIKGLCQSGKLEDARKVFEGLLVKGHNLNV 1083
Query: 446 SSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFLPEPKTESML 486
+Y ++I G + EA +M++ G +P+ KT ++
Sbjct: 1084DTYTIMIQGFCVEGLFNEALALLSKMEDNGCIPDAKTYEII 1206
Score = 73.2 bits (178), Expect = 2e-13
Identities = 41/146 (28%), Positives = 74/146 (50%)
Frame = +2
Query: 236 DTKSFNIILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQL 295
DT +F + G C L + A +++ D +SY ++I K + L L
Sbjct: 26 DTITFTTLSKGLC-LKGQIQQAFLFHDKVVALGFHFDQISYGTLIHGLCKVGETRAALDL 202
Query: 296 FEQMKKRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCK 355
+++ + P+ +YN +I S+ K ++V EA +L +M ++PD +TY++LI C
Sbjct: 203 LQRVDGNLVQPNVVMYNTIIDSMCKVKLVNEAFDLFSEMVSKGISPDVVTYSALISGFCI 382
Query: 356 ARKIDEAKEIFNVMLERGISPSIRTF 381
K+ +A ++FN M+ I P + TF
Sbjct: 383 LGKLKDAIDLFNKMILENIKPDVYTF 460
Score = 63.9 bits (154), Expect = 2e-10
Identities = 33/110 (30%), Positives = 63/110 (57%), Gaps = 3/110 (2%)
Frame = +2
Query: 307 DRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIF 366
D+ Y +I L K + A++L+ +++ N V P+ + YN++I +CK + ++EA ++F
Sbjct: 131 DQISYGTLIHGLCKVGETRAALDLLQRVDGNLVQPNVVMYNTIIDSMCKVKLVNEAFDLF 310
Query: 367 NVMLERGISPSIRTFHAF---FRILRVKEEVFELLDKMKELGCNPTIETY 413
+ M+ +GISP + T+ A F IL ++ +L +KM P + T+
Sbjct: 311 SEMVSKGISPDVVTYSALISGFCILGKLKDAIDLFNKMILENIKPDVYTF 460
>CA991373 weakly similar to PIR|D86260|D86 protein T12C24.22 [imported] -
Arabidopsis thaliana, partial (6%)
Length = 756
Score = 118 bits (295), Expect = 7e-27
Identities = 71/230 (30%), Positives = 125/230 (53%), Gaps = 6/230 (2%)
Frame = +2
Query: 209 LLSALCRYKNVQDAEHLLFC--NKNVFPLDTKSFNIILNGWCNLIVSARNAERIWEEMSK 266
L+ C K V A+ +L+ + V P D +S+NI+++G+C I A +++EM
Sbjct: 44 LMDGYCLVKEVNKAKSILYTMSQRGVNP-DIQSYNILIDGFCK-IKKVDEAMNLFKEMHH 217
Query: 267 RRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYNAVIFSLAKNRMVKE 326
+ I DVV+Y S+I K K+ L+L ++M R + PD Y++++ +L KN V +
Sbjct: 218 KHIIPDVVTYNSLIDGLCKLGKISYALKLVDEMHDRGVPPDIITYSSILDALCKNHQVDK 397
Query: 327 AVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRTF----H 382
A+ L+ K++D + P+ TY LI LCK ++++A IF +L +G + ++ T+ H
Sbjct: 398 AIALLTKLKDQGIRPNMYTYTILIDGLCKGGRLEDAHNIFEDLLVKGYNITVNTYTVMIH 577
Query: 383 AFFRILRVKEEVFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRI 432
F + +E LL KMK+ C P TY ++IR + D+ +++
Sbjct: 578 GFCN-KGLFDEALTLLSKMKDNSCIPDAVTYEIIIRSLFDKDENDKAEKL 724
Score = 115 bits (287), Expect = 6e-26
Identities = 67/224 (29%), Positives = 115/224 (50%), Gaps = 3/224 (1%)
Frame = +2
Query: 266 KRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYNAVIFSLAKNRMVK 325
K+ I+ VV+Y S++ Y ++ + + M +R + PD + YN +I K + V
Sbjct: 5 KQGIKPIVVTYCSLMDGYCLVKEVNKAKSILYTMSQRGVNPDIQSYNILIDGFCKIKKVD 184
Query: 326 EAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRTFHAFF 385
EA+NL +M ++ PD +TYNSLI LCK KI A ++ + M +RG+ P I T+ +
Sbjct: 185 EAMNLFKEMHHKHIIPDVVTYNSLIDGLCKLGKISYALKLVDEMHDRGVPPDIITYSSIL 364
Query: 386 RILRVKEEV---FELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDGIG 442
L +V LL K+K+ G P + TY +LI C+ +L++ I+ + G
Sbjct: 365 DALCKNHQVDKAIALLTKLKDQGIRPNMYTYTILIDGLCKGGRLEDAHNIFEDLLVKGYN 544
Query: 443 HDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFLPEPKTESML 486
++Y V+IHG +EA +M++ +P+ T ++
Sbjct: 545 ITVNTYTVMIHGFCNKGLFDEALTLLSKMKDNSCIPDAVTYEII 676
Score = 113 bits (282), Expect = 2e-25
Identities = 69/238 (28%), Positives = 124/238 (51%), Gaps = 3/238 (1%)
Frame = +2
Query: 243 ILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKR 302
+++G+C L+ A+ I MS+R + D+ SY +I + K K+ + LF++M +
Sbjct: 44 LMDGYC-LVKEVNKAKSILYTMSQRGVNPDIQSYNILIDGFCKIKKVDEAMNLFKEMHHK 220
Query: 303 NITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEA 362
+I PD YN++I L K + A+ L+ +M D V PD ITY+S++ LCK ++D+A
Sbjct: 221 HIIPDVVTYNSLIDGLCKLGKISYALKLVDEMHDRGVPPDIITYSSILDALCKNHQVDKA 400
Query: 363 KEIFNVMLERGISPSIRTFHAFFRIL---RVKEEVFELLDKMKELGCNPTIETYIMLIRK 419
+ + ++GI P++ T+ L E+ + + + G N T+ TY ++I
Sbjct: 401 IALLTKLKDQGIRPNMYTYTILIDGLCKGGRLEDAHNIFEDLLVKGYNITVNTYTVMIHG 580
Query: 420 FCRWRQLDEVKRIWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFL 477
FC DE + + M+++ D +Y ++I LF + ++A K EM +G L
Sbjct: 581 FCNKGLFDEALTLLSKMKDNSCIPDAVTYEIIIRSLFDKDENDKAEKLR-EMITRGLL 751
>BF004907 weakly similar to GP|8953393|emb putative protein {Arabidopsis
thaliana}, partial (34%)
Length = 665
Score = 111 bits (277), Expect = 8e-25
Identities = 65/214 (30%), Positives = 112/214 (51%), Gaps = 3/214 (1%)
Frame = +3
Query: 269 IQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYNAVIFSLAKNRMVKEAV 328
++ VV+Y +++ Y + ++ + L++ +M K I P+ VYN +I +LA+ KEA+
Sbjct: 15 VRPSVVTYGTLVEGYCRMRRVEKALEMVGEMTKEGIKPNAIVYNPIIDALAEAGRFKEAL 194
Query: 329 NLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRTFHAFFRIL 388
++ + + P TYNSL+K CKA I+ A +I M+ RG P T++ FFR
Sbjct: 195 GMMERFHVLQIGPTLSTYNSLVKGFCKAGDIEGASKILKKMISRGFLPIPTTYNYFFRYF 374
Query: 389 R---VKEEVFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDGIGHDR 445
+E L KM E G NP TY ++++ C +L+ ++ MR G D
Sbjct: 375 SRCGKVDEGMNLYTKMIESGHNPDRLTYHLVLKMLCEEEKLELAVQVSMEMRHKGYDMDL 554
Query: 446 SSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFLPE 479
++ +L H L K+EEA+ + +M +G +P+
Sbjct: 555 ATSTMLTHLLCKMHKLEEAFAEFEDMIRRGIIPQ 656
Score = 76.3 bits (186), Expect = 3e-14
Identities = 52/219 (23%), Positives = 100/219 (44%), Gaps = 6/219 (2%)
Frame = +3
Query: 229 NKNVFPLDTKSFNIILNGWCNLIVSARNAERIWE---EMSKRRIQHDVVSYASIISCYSK 285
N+NV P ++ ++ G+C + R E+ E EM+K I+ + + Y II ++
Sbjct: 6 NENVRP-SVVTYGTLVEGYCRM----RRVEKALEMVGEMTKEGIKPNAIVYNPIIDALAE 170
Query: 286 SSKLYRVLQLFEQMKKRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAIT 345
+ + L + E+ I P YN+++ K ++ A ++ KM P T
Sbjct: 171 AGRFKEALGMMERFHVLQIGPTLSTYNSLVKGFCKAGDIEGASKILKKMISRGFLPIPTT 350
Query: 346 YNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRTFHAFFRILRVKEEV---FELLDKMK 402
YN + + K+DE ++ M+E G +P T+H ++L +E++ ++ +M+
Sbjct: 351 YNYFFRYFSRCGKVDEGMNLYTKMIESGHNPDRLTYHLVLKMLCEEEKLELAVQVSMEMR 530
Query: 403 ELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDGI 441
G + + T ML C+ +L+E + M GI
Sbjct: 531 HKGYDMDLATSTMLTHLLCKMHKLEEAFAEFEDMIRRGI 647
Score = 60.8 bits (146), Expect = 1e-09
Identities = 51/219 (23%), Positives = 93/219 (42%), Gaps = 2/219 (0%)
Frame = +3
Query: 164 SIVTPQTLLIMIRKYCAVHDVGRAINTFYAFKRFNFQVGLYEFQGLLSALCRYKNVQDAE 223
S+VT TL + YC + V +A+ + + + ++ AL ++A
Sbjct: 24 SVVTYGTL---VEGYCRMRRVEKALEMVGEMTKEGIKPNAIVYNPIIDALAEAGRFKEAL 194
Query: 224 HLL--FCNKNVFPLDTKSFNIILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIIS 281
++ F + P ++N ++ G+C A +I ++M R +Y
Sbjct: 195 GMMERFHVLQIGPT-LSTYNSLVKGFCKA-GDIEGASKILKKMISRGFLPIPTTYNYFFR 368
Query: 282 CYSKSSKLYRVLQLFEQMKKRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTP 341
+S+ K+ + L+ +M + PDR Y+ V+ L + ++ AV + ++M
Sbjct: 369 YFSRCGKVDEGMNLYTKMIESGHNPDRLTYHLVLKMLCEEEKLELAVQVSMEMRHKGYDM 548
Query: 342 DAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRT 380
D T L LCK K++EA F M+ RGI P T
Sbjct: 549 DLATSTMLTHLLCKMHKLEEAFAEFEDMIRRGIIPQYLT 665
Score = 53.5 bits (127), Expect = 2e-07
Identities = 44/215 (20%), Positives = 94/215 (43%), Gaps = 7/215 (3%)
Frame = +3
Query: 195 KRFNFQVGLYEFQGLLSALCRYKNVQDAEHLLFCNKNVFPLDTKSFNIILNGWCNLIVSA 254
K N + + + L+ CR + V+ A ++ + K I+ N + + A
Sbjct: 3 KNENVRPSVVTYGTLVEGYCRMRRVEKALEMV---GEMTKEGIKPNAIVYNPIIDALAEA 173
Query: 255 ---RNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVY 311
+ A + E +I + +Y S++ + K+ + ++ ++M R P Y
Sbjct: 174 GRFKEALGMMERFHVLQIGPTLSTYNSLVKGFCKAGDIEGASKILKKMISRGFLPIPTTY 353
Query: 312 NAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLE 371
N ++ V E +NL KM ++ PD +TY+ ++K LC+ K++ A ++ M
Sbjct: 354 NYFFRYFSRCGKVDEGMNLYTKMIESGHNPDRLTYHLVLKMLCEEEKLELAVQVSMEMRH 533
Query: 372 RG----ISPSIRTFHAFFRILRVKEEVFELLDKMK 402
+G ++ S H ++ +++E E D ++
Sbjct: 534 KGYDMDLATSTMLTHLLCKMHKLEEAFAEFEDMIR 638
>TC84326 weakly similar to PIR|F85225|F85225 hypothetical protein AT4g19900
[imported] - Arabidopsis thaliana, partial (18%)
Length = 756
Score = 104 bits (260), Expect = 8e-23
Identities = 70/244 (28%), Positives = 116/244 (46%), Gaps = 5/244 (2%)
Frame = +1
Query: 240 FNIILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQM 299
+ ++ G+C R AE + M ++ + + +Y ++I + K+ R L M
Sbjct: 1 YTAMIRGYCREDKLNR-AEMLLSRMKEQGLVPNTNTYTTLIDGHCKAGNFERAYDLMNLM 177
Query: 300 KKRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKI 359
+P+ YNA++ L K V+EA ++ N + PD TYN L+ CK I
Sbjct: 178 SSEGFSPNLCTYNAIVNGLCKRGRVQEAYKMLEDGFQNGLKPDKFTYNILMSEHCKQENI 357
Query: 360 DEAKEIFNVMLERGISPSIRTF----HAFFRILRVKE-EVFELLDKMKELGCNPTIETYI 414
+A +FN ML+ GI P I ++ F R R+KE E+F ++ +G PT +TY
Sbjct: 358 RQALALFNKMLKIGIQPDIHSYTTLIAVFCRENRMKESEMF--FEEAVRIGIIPTNKTYT 531
Query: 415 MLIRKFCRWRQLDEVKRIWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEK 474
+I +CR L + ++ + + G + +Y +I GL K +EA Y M EK
Sbjct: 532 SMICGYCREGNLTLAMKFFHRLSDHGCAPESITYGAIISGLCKQSKRDEARSLYDSMIEK 711
Query: 475 GFLP 478
G +P
Sbjct: 712 GLVP 723
Score = 92.0 bits (227), Expect = 5e-19
Identities = 55/222 (24%), Positives = 108/222 (47%), Gaps = 1/222 (0%)
Frame = +1
Query: 156 MRRGKTGESIVTPQTLLIMIRKYCAVHDVGRAINTFYAFKRFNFQVGLYEFQGLLSALCR 215
+ R K + T +I +C + RA + F L + +++ LC+
Sbjct: 61 LSRMKEQGLVPNTNTYTTLIDGHCKAGNFERAYDLMNLMSSEGFSPNLCTYNAIVNGLCK 240
Query: 216 YKNVQDAEHLLFCN-KNVFPLDTKSFNIILNGWCNLIVSARNAERIWEEMSKRRIQHDVV 274
VQ+A +L +N D ++NI+++ C + R A ++ +M K IQ D+
Sbjct: 241 RGRVQEAYKMLEDGFQNGLKPDKFTYNILMSEHCKQ-ENIRQALALFNKMLKIGIQPDIH 417
Query: 275 SYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKM 334
SY ++I+ + + +++ FE+ + I P K Y ++I + + A+ ++
Sbjct: 418 SYTTLIAVFCRENRMKESEMFFEEAVRIGIIPTNKTYTSMICGYCREGNLTLAMKFFHRL 597
Query: 335 EDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISP 376
D+ P++ITY ++I LCK K DEA+ +++ M+E+G+ P
Sbjct: 598 SDHGCAPESITYGAIISGLCKQSKRDEARSLYDSMIEKGLVP 723
>BQ148309 weakly similar to GP|6721172|gb| hypothetical protein {Arabidopsis
thaliana}, partial (30%)
Length = 654
Score = 92.4 bits (228), Expect = 4e-19
Identities = 45/159 (28%), Positives = 81/159 (50%)
Frame = +1
Query: 92 VKASSELVVEVLSRVRNDWEAAFTFFLWAGKQPGYDHSVREYHSMISVLGKMRRFDTAWA 151
+ S L+ +L R ++DW++A F WAG + HS + Y M+ +LG+M+ D
Sbjct: 181 IHLSENLINRLLFRYKDDWKSALAIFRWAGSHSNFKHSQQSYDMMVDILGRMKAMDKMRE 360
Query: 152 LVEEMRRGKTGESIVTPQTLLIMIRKYCAVHDVGRAINTFYAFKRFNFQVGLYEFQGLLS 211
++EEMR+ ES++T T+ ++R++ A+ F + + LL
Sbjct: 361 ILEEMRQ----ESLITLNTIAKVMRRFVGARQWKDAVRIFDDLQFLGLEKNTESMNVLLD 528
Query: 212 ALCRYKNVQDAEHLLFCNKNVFPLDTKSFNIILNGWCNL 250
LC+ K V+ A + K+ + +FNI+++GWCN+
Sbjct: 529 TLCKEKFVEQAREIYLELKHYIAPNAHTFNILIHGWCNI 645
>TC81904 weakly similar to GP|8778410|gb|AAF79418.1| F16A14.3 {Arabidopsis
thaliana}, partial (7%)
Length = 978
Score = 89.4 bits (220), Expect = 3e-18
Identities = 66/288 (22%), Positives = 124/288 (42%), Gaps = 11/288 (3%)
Frame = +1
Query: 174 MIRKYCAVHDVGRAINTFYAFKRFNFQVGLYEFQGLLSALCRYKNVQDAEHLL--FCNKN 231
++ YC + +A+ + + + F +L L + + F
Sbjct: 85 LVHGYCNSRNFDKALAVYKSMISRGIKTNCVIFSCILHCLDEMGRALEVVDMFEEFKESG 264
Query: 232 VFPLDTKSFNIILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYR 291
+F +D K++NI+ + C L +A + +E+ ++ D+ Y ++I+ Y K
Sbjct: 265 LF-IDRKAYNILFDALCKL-GKVDDAVGMLDELKSMQLDVDMKHYTTLINGYFLQGKPIE 438
Query: 292 VLQLFEQMKKRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIK 351
LF++M++R PD YN + +NR EA++L+ ME V P++ T+ +I+
Sbjct: 439 AQSLFKEMEERGFKPDVVAYNVLAAGFFRNRTDFEAMDLLNYMESQGVEPNSTTHKIIIE 618
Query: 352 PLCKARKIDEAKEIFNVMLERGISPSIRTFHAFFR-------ILRVKE--EVFELLDKMK 402
LC A K++EA+E FN + + S+ + A I + E E F LL M
Sbjct: 619 GLCSAGKVEEAEEFFNWLKGESVEISVEIYTALVNGYCEAALIEKSHELKEAFILLRTML 798
Query: 403 ELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDGIGHDRSSYIV 450
E+ P+ Y + C ++ ++N G D +Y +
Sbjct: 799 EMNMKPSKVMYSKIFTALCCNGNMEGAHTLFNLFXHTGFTPDAVTYTI 942
Score = 80.9 bits (198), Expect = 1e-15
Identities = 54/192 (28%), Positives = 93/192 (48%), Gaps = 3/192 (1%)
Frame = +1
Query: 326 EAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRTFHAFF 385
EA ++ ++ME + PD Y +L+ C +R D+A ++ M+ RGI + F
Sbjct: 16 EAESVFLEMEKQGLVPDVYVYCALVHGYCNSRNFDKALAVYKSMISRGIKTNCVIFSCIL 195
Query: 386 RILRVKE---EVFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDGIG 442
L EV ++ ++ KE G + Y +L C+ ++D+ + + ++ +
Sbjct: 196 HCLDEMGRALEVVDMFEEFKESGLFIDRKAYNILFDALCKLGKVDDAVGMLDELKSMQLD 375
Query: 443 HDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFLPEPKTESMLQAWLSGRQVTDSQATD 502
D Y LI+G FL K EA + EM+E+GF P+ ++L A R TD +A D
Sbjct: 376 VDMKHYTTLINGYFLQGKPIEAQSLFKEMEERGFKPDVVAYNVLAAGFF-RNRTDFEAMD 552
Query: 503 LEHNQLEHGGLK 514
L N +E G++
Sbjct: 553 L-LNYMESQGVE 585
Score = 77.4 bits (189), Expect = 1e-14
Identities = 55/217 (25%), Positives = 105/217 (48%), Gaps = 5/217 (2%)
Frame = +1
Query: 257 AERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYNAVIF 316
AE ++ EM K+ + DV Y +++ Y S + L +++ M R I + +++ ++
Sbjct: 19 AESVFLEMEKQGLVPDVYVYCALVHGYCNSRNFDKALAVYKSMISRGIKTNCVIFSCILH 198
Query: 317 SLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEA----KEIFNVMLER 372
L + E V++ + +++ + D YN L LCK K+D+A E+ ++ L+
Sbjct: 199 CLDEMGRALEVVDMFEEFKESGLFIDRKAYNILFDALCKLGKVDDAVGMLDELKSMQLDV 378
Query: 373 GISPSIRTFHAFFRILRVKE-EVFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKR 431
+ + +F L+ K E L +M+E G P + Y +L F R R E
Sbjct: 379 DMKHYTTLINGYF--LQGKPIEAQSLFKEMEERGFKPDVVAYNVLAAGFFRNRTDFEAMD 552
Query: 432 IWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAYKYY 468
+ N M G+ + +++ ++I GL KVEEA +++
Sbjct: 553 LLNYMESQGVEPNSTTHKIIIEGLCSAGKVEEAEEFF 663
>TC89807 weakly similar to PIR|B96600|B96600 protein F14J16.14 [imported] -
Arabidopsis thaliana, partial (53%)
Length = 1241
Score = 88.6 bits (218), Expect = 6e-18
Identities = 71/279 (25%), Positives = 143/279 (50%), Gaps = 9/279 (3%)
Frame = +1
Query: 163 ESIVTPQTLLIMIRKYCAVHDVGRAINTFYAFKRFNFQVGLYEFQ-GLLSALCRYKNVQD 221
+ + Q L ++ K+ D+ R F++ N G+YE L+ R++ V+D
Sbjct: 73 QDLYKEQNLKTLVEKFKKASDIDR-------FRKKN---GIYEDTVRRLAGAKRFRWVRD 222
Query: 222 A-EHLLFCNKNVFPLDTKSFN---IILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYA 277
EH K+ + + F+ I L G N+ R+A+++++EM +R + V+S
Sbjct: 223 IIEH----QKSYADISNEGFSARLITLYGKSNM---HRHAQKLFDEMPQRNCERSVLSLN 381
Query: 278 SIISCYSKSSKLYRVLQLFEQMKKR-NITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMED 336
++++ Y S + V +LF+++ + ++ PD YN I +L + AV+++ +ME
Sbjct: 382 ALLAAYLHSKQYDVVERLFKKLPVQLSVKPDLVSYNTYIKALLEKGSFDSAVSVLEEMEK 561
Query: 337 NNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRTFHAFFRILRVKE---E 393
+ V D IT+N+L+ L + ++ ++++ + E+ + P+IRT++A L V + E
Sbjct: 562 DGVESDLITFNTLLDGLYSKGRFEDGEKLWEKLGEKNVVPNIRTYNARLLGLAVAKRAGE 741
Query: 394 VFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRI 432
E ++M++ G P + ++ LI+ F LDE K +
Sbjct: 742 AVEFYEEMEKKGVKPDLFSFNALIKGFANEGNLDEAKNV 858
Score = 42.0 bits (97), Expect = 6e-04
Identities = 41/163 (25%), Positives = 73/163 (44%), Gaps = 5/163 (3%)
Frame = +1
Query: 362 AKEIFNVMLERGISPSIRTFHAFFRI-LRVKE-EVFELLDKMK--ELGCNPTIETYIMLI 417
A+++F+ M +R S+ + +A L K+ +V E L K +L P + +Y I
Sbjct: 319 AQKLFDEMPQRNCERSVLSLNALLAAYLHSKQYDVVERLFKKLPVQLSVKPDLVSYNTYI 498
Query: 418 RKFCRWRQLDEVKRIWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFL 477
+ D + M +DG+ D ++ L+ GL+ + E+ K + ++ EK +
Sbjct: 499 KALLEKGSFDSAVSVLEEMEKDGVESDLITFNTLLDGLYSKGRFEDGEKLWEKLGEKNVV 678
Query: 478 PEPKTESMLQAWLSGRQVTDSQATDLE-HNQLEHGGLKKSVKP 519
P +T A L G V +E + ++E KK VKP
Sbjct: 679 PNIRT---YNARLLGLAVAKRAGEAVEFYEEME----KKGVKP 786
>BF003426 similar to PIR|F96760|F96 hypothetical protein T9L24.39 [imported]
- Arabidopsis thaliana, partial (43%)
Length = 774
Score = 88.2 bits (217), Expect = 7e-18
Identities = 55/157 (35%), Positives = 80/157 (50%), Gaps = 4/157 (2%)
Frame = +2
Query: 334 MEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRTFHAFFRIL---RV 390
M + PD TY +I+ +C KIDEA + M ++G P I T + F ++L +
Sbjct: 20 MISSGCLPDVTTYKDIIEGMCLCGKIDEAYKFLEEMGKKGYPPDIVTHNCFLKVLCHNKK 199
Query: 391 KEEVFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDGIGHDRSSYIV 450
EE +L +M EL C P+++TY MLI F + D W+ M + G D +Y V
Sbjct: 200 SEEALKLYGRMIELSCIPSVQTYNMLISMFFKMDDPDGAFETWHEMEKRGCRPDTDTYGV 379
Query: 451 LIHGLFLNCKVEEAYKYYIEMQEKGF-LPEPKTESML 486
+I GLF K E+A E+ KG LP K +S+L
Sbjct: 380 MIEGLFNCNKAEDACILLEEVINKGIKLPYRKFDSLL 490
Score = 53.9 bits (128), Expect = 2e-07
Identities = 42/183 (22%), Positives = 76/183 (40%), Gaps = 3/183 (1%)
Frame = +2
Query: 236 DTKSFNIILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQL 295
D ++ I+ G C L A + EEM K+ D+V++ + + K L+L
Sbjct: 44 DVTTYKDIIEGMC-LCGKIDEAYKFLEEMGKKGYPPDIVTHNCFLKVLCHNKKSEEALKL 220
Query: 296 FEQMKKRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCK 355
+ +M + + P + YN +I K A +ME PD TY +I+ L
Sbjct: 221 YGRMIELSCIPSVQTYNMLISMFFKMDDPDGAFETWHEMEKRGCRPDTDTYGVMIEGLFN 400
Query: 356 ARKIDEAKEIFNVMLERGISPSIRTFHAFFRILRV---KEEVFELLDKMKELGCNPTIET 412
K ++A + ++ +GI R F + L + + +L D M++ NP
Sbjct: 401 CNKAEDACILLEEVINKGIKLPYRKFDSLLMQLSEIGNLQAIHKLSDHMRKFYNNPMARR 580
Query: 413 YIM 415
+ +
Sbjct: 581 FAL 589
Score = 43.9 bits (102), Expect = 2e-04
Identities = 28/92 (30%), Positives = 42/92 (45%)
Frame = +2
Query: 395 FELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDGIGHDRSSYIVLIHG 454
F+ + M GC P + TY +I C ++DE + M + G D ++ +
Sbjct: 2 FKFMGHMISSGCLPDVTTYKDIIEGMCLCGKIDEAYKFLEEMGKKGYPPDIVTHNCFLKV 181
Query: 455 LFLNCKVEEAYKYYIEMQEKGFLPEPKTESML 486
L N K EEA K Y M E +P +T +ML
Sbjct: 182 LCHNKKSEEALKLYGRMIELSCIPSVQTYNML 277
>AW774457 weakly similar to GP|10176973|dbj
gb|AAF19552.1~gene_id:MTG13.9~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (13%)
Length = 650
Score = 87.8 bits (216), Expect = 1e-17
Identities = 62/235 (26%), Positives = 107/235 (45%)
Frame = -1
Query: 243 ILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKR 302
++NG+C I A ++ +M + I D V+Y S+I KS ++ +L ++M
Sbjct: 650 MINGFCK-IKMVDEALSLFNDMQFKGIAPDKVTYNSLIDGLCKSGRISYAWELVDEMHDN 474
Query: 303 NITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEA 362
+ YN +I +L KN V +A+ L+ K++D + PD T+N LI LCK ++ A
Sbjct: 473 GQPANIFTYNCLIDALCKNHHVDQAIALVKKIKDQGIQPDMYTFNILIYGLCKVGRLKNA 294
Query: 363 KEIFNVMLERGISPSIRTFHAFFRILRVKEEVFELLDKMKELGCNPTIETYIMLIRKFCR 422
+++F +L +G S + TY +++ C+
Sbjct: 293 QDVFQDLLSKGYSVN--------------------------------AWTYNIMVNGLCK 210
Query: 423 WRQLDEVKRIWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFL 477
DE + + + M ++GI D +Y LI LF + E+A K EM +G L
Sbjct: 209 EGLFDEAEALLSKMDDNGIIPDAVTYETLIQALFHKDENEKAEKLLREMIARGLL 45
>BE998743 similar to GP|22128591|gb| fertility restorer-like protein {Petunia
x hybrida}, partial (7%)
Length = 829
Score = 86.7 bits (213), Expect = 2e-17
Identities = 60/234 (25%), Positives = 111/234 (46%), Gaps = 7/234 (2%)
Frame = +2
Query: 253 SARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYN 312
+ + A+ M K I+ DVV+Y S++ Y +++ + + + + + PD + YN
Sbjct: 116 NVKGAKNALAMMIKGSIKPDVVTYNSLMDGYCLVNEVNKAKHVLSTIARMGVAPDAQSYN 295
Query: 313 AVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLER 372
++ + + A L+ +M N PD TY+SLI LCK +D+A + + ++
Sbjct: 296 IMVNGFCR---ISHAWKLVDEMHVNGQPPDIFTYSSLIDALCKNNHLDKAIALVKKIKDQ 466
Query: 373 GISPSIRTFHAFF-------RILRVKEEVFELLDKMKELGCNPTIETYIMLIRKFCRWRQ 425
GI P++ T++ R+ ++ +LL K G + I TY +LI C+
Sbjct: 467 GIQPNMYTYNILIDGLCKGGRLKNAQDVFQDLLTK----GYSLNIRTYNILINGLCKEGL 634
Query: 426 LDEVKRIWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFLPE 479
D+ + + + M ++ I + +Y +I LF E+A K EM +G L E
Sbjct: 635 FDKAEALLSKMEDNDINPNVVTYETIIRSLFYKDYNEKAEKLLREMVARGLL*E 796
Score = 85.5 bits (210), Expect = 5e-17
Identities = 57/205 (27%), Positives = 103/205 (49%), Gaps = 5/205 (2%)
Frame = +2
Query: 206 FQGLLSALCRYKNVQDAEHLL--FCNKNVFPLDTKSFNIILNGWCNLIVSARNAERIWEE 263
+ L+ C V A+H+L V P D +S+NI++NG+C + +A ++ +E
Sbjct: 185 YNSLMDGYCLVNEVNKAKHVLSTIARMGVAP-DAQSYNIMVNGFCRI----SHAWKLVDE 349
Query: 264 MSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYNAVIFSLAKNRM 323
M D+ +Y+S+I K++ L + + L +++K + I P+ YN +I L K
Sbjct: 350 MHVNGQPPDIFTYSSLIDALCKNNHLDKAIALVKKIKDQGIQPNMYTYNILIDGLCKGGR 529
Query: 324 VKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRTFHA 383
+K A ++ + + + TYN LI LCK D+A+ + + M + I+P++ T+
Sbjct: 530 LKNAQDVFQDLLTKGYSLNIRTYNILINGLCKEGLFDKAEALLSKMEDNDINPNVVTYET 709
Query: 384 FFRILRVK---EEVFELLDKMKELG 405
R L K E+ +LL +M G
Sbjct: 710 IIRSLFYKDYNEKAEKLLREMVARG 784
Score = 77.0 bits (188), Expect = 2e-14
Identities = 43/169 (25%), Positives = 85/169 (49%)
Frame = +2
Query: 318 LAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPS 377
+ K VK A N + M ++ PD +TYNSL+ C ++++AK + + + G++P
Sbjct: 101 MRKEGNVKGAKNALAMMIKGSIKPDVVTYNSLMDGYCLVNEVNKAKHVLSTIARMGVAPD 280
Query: 378 IRTFHAFFRILRVKEEVFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMR 437
++++ ++L+D+M G P I TY LI C+ LD+ + ++
Sbjct: 281 AQSYNIMVNGFCRISHAWKLVDEMHVNGQPPDIFTYSSLIDALCKNNHLDKAIALVKKIK 460
Query: 438 EDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFLPEPKTESML 486
+ GI + +Y +LI GL +++ A + ++ KG+ +T ++L
Sbjct: 461 DQGIQPNMYTYNILIDGLCKGGRLKNAQDVFQDLLTKGYSLNIRTYNIL 607
>BF632209 weakly similar to GP|8493579|gb|A Contains a RepB PF|01051 protein
domain and multiple PPR PF|01535 repeats. EST
gb|AA728420 comes, partial (13%)
Length = 508
Score = 81.6 bits (200), Expect = 7e-16
Identities = 45/132 (34%), Positives = 77/132 (58%)
Frame = +1
Query: 236 DTKSFNIILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQL 295
+ +S++I++NG+C I A ++ EM RRI D V+Y S+I KS ++ +L
Sbjct: 112 NARSYSIVINGFCK-IKMVDKALSLFYEMRCRRIAPDTVTYNSLIDGLCKSGRISYAWEL 288
Query: 296 FEQMKKRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCK 355
++M+ D YN++I +L KN V +A+ L+ K++D + D TYN LI LCK
Sbjct: 289 VDEMRDSGQPADIITYNSLIDALCKNHHVDKAIALVKKIKDQGIQLDMYTYNILIDGLCK 468
Query: 356 ARKIDEAKEIFN 367
++++A+ IF+
Sbjct: 469 QGRLNDAQVIFH 504
Score = 74.3 bits (181), Expect = 1e-13
Identities = 39/119 (32%), Positives = 67/119 (55%)
Frame = +1
Query: 264 MSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYNAVIFSLAKNRM 323
+S+ R+ + SY+ +I+ + K + + L LF +M+ R I PD YN++I L K+
Sbjct: 88 ISRMRVAPNARSYSIVINGFCKIKMVDKALSLFYEMRCRRIAPDTVTYNSLIDGLCKSGR 267
Query: 324 VKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRTFH 382
+ A L+ +M D+ D ITYNSLI LCK +D+A + + ++GI + T++
Sbjct: 268 ISYAWELVDEMRDSGQPADIITYNSLIDALCKNHHVDKAIALVKKIKDQGIQLDMYTYN 444
Score = 70.5 bits (171), Expect = 2e-12
Identities = 41/139 (29%), Positives = 74/139 (52%), Gaps = 3/139 (2%)
Frame = +1
Query: 299 MKKRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARK 358
+ + + P+ + Y+ VI K +MV +A++L +M + PD +TYNSLI LCK+ +
Sbjct: 88 ISRMRVAPNARSYSIVINGFCKIKMVDKALSLFYEMRCRRIAPDTVTYNSLIDGLCKSGR 267
Query: 359 IDEAKEIFNVMLERGISPSIRTFHAFFRILRVKEEV---FELLDKMKELGCNPTIETYIM 415
I A E+ + M + G I T+++ L V L+ K+K+ G + TY +
Sbjct: 268 ISYAWELVDEMRDSGQPADIITYNSLIDALCKNHHVDKAIALVKKIKDQGIQLDMYTYNI 447
Query: 416 LIRKFCRWRQLDEVKRIWN 434
LI C+ +L++ + I++
Sbjct: 448 LIDGLCKQGRLNDAQVIFH 504
Score = 48.1 bits (113), Expect = 9e-06
Identities = 30/91 (32%), Positives = 43/91 (46%)
Frame = +1
Query: 133 YHSMISVLGKMRRFDTAWALVEEMRRGKTGESIVTPQTLLIMIRKYCAVHDVGRAINTFY 192
Y+S+I L K R AW LV+EMR I+T +L I C H V +AI
Sbjct: 229 YNSLIDGLCKSGRISYAWELVDEMRDSGQPADIITYNSL---IDALCKNHHVDKAIALVK 399
Query: 193 AFKRFNFQVGLYEFQGLLSALCRYKNVQDAE 223
K Q+ +Y + L+ LC+ + DA+
Sbjct: 400 KIKDQGIQLDMYTYNILIDGLCKQGRLNDAQ 492
Score = 29.6 bits (65), Expect = 3.1
Identities = 9/29 (31%), Positives = 20/29 (68%)
Frame = +2
Query: 341 PDAITYNSLIKPLCKARKIDEAKEIFNVM 369
PD +TY+SL+ C ++++AK + +++
Sbjct: 2 PDVVTYSSLMDGYCLVNEVNKAKHVLSIL 88
>TC93383 similar to PIR|T47786|T47786 hypothetical protein F17J16.90 -
Arabidopsis thaliana, partial (43%)
Length = 1038
Score = 79.7 bits (195), Expect = 3e-15
Identities = 56/274 (20%), Positives = 121/274 (43%)
Frame = +1
Query: 242 IILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKK 301
+ N + + + I+++M + ++ DVVSYA +I+ Y K+ + L +FE+M
Sbjct: 7 VTYNSLMSFETNYKEVSNIYDQMQRADLRPDVVSYALLINAYGKARREEEALAVFEEMLD 186
Query: 302 RNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDE 361
+ P RK YN ++ + + + MV++A + M + PD +Y +++ A ++
Sbjct: 187 AGVRPTRKAYNILLDAFSISGMVEQARIVFKSMRRDKYMPDLCSYTTMLSAYVNAPDMEG 366
Query: 362 AKEIFNVMLERGISPSIRTFHAFFRILRVKEEVFELLDKMKELGCNPTIETYIMLIRKFC 421
A++ F +++ G P+ + TY LI+ +
Sbjct: 367 AEKFFKRLIQDGFEPN--------------------------------VVTYGTLIKGYA 450
Query: 422 RWRQLDEVKRIWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFLPEPK 481
+ +++V + M GI +++ ++ N + A ++ EM G LP+ K
Sbjct: 451 KANDIEKVMEKYEEMLGRGIKANQTILTTIMDAHGKNGDFDSAVNWFKEMALNGLLPDQK 630
Query: 482 TESMLQAWLSGRQVTDSQATDLEHNQLEHGGLKK 515
+++L + L+ + +A +L + +E L K
Sbjct: 631 AKNILLS-LAKTEEDIKEANELVLHSIEINNLPK 729
Score = 50.4 bits (119), Expect = 2e-06
Identities = 52/216 (24%), Positives = 99/216 (45%), Gaps = 6/216 (2%)
Frame = +1
Query: 130 VREYHSMISVLGKMRRFDTAWALVEEMRRGKTGESIVTPQTLLIMIRKYCAVHDVGRAIN 189
V Y +I+ GK RR + A A+ EEM T + I++ + V +A
Sbjct: 100 VVSYALLINAYGKARREEEALAVFEEMLDAGVRP---TRKAYNILLDAFSISGMVEQARI 270
Query: 190 TFYAFKRFNFQVGLYEFQGLLSALCRYKNVQDAEHLLF-CNKNVFPLDTKSFNIILNGWC 248
F + +R + L + +LSA +++ AE ++ F + ++ ++ G+
Sbjct: 271 VFKSMRRDKYMPDLCSYTTMLSAYVNAPDMEGAEKFFKRLIQDGFEPNVVTYGTLIKGYA 450
Query: 249 NLIVSARNAERI---WEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNIT 305
A + E++ +EEM R I+ + +I+ + K+ + F++M +
Sbjct: 451 K----ANDIEKVMEKYEEMLGRGIKANQTILTTIMDAHGKNGDFDSAVNWFKEMALNGLL 618
Query: 306 PDRKVYNAVIFSLAKNRM-VKEAVNLII-KMEDNNV 339
PD+K N ++ SLAK +KEA L++ +E NN+
Sbjct: 619 PDQKAKN-ILLSLAKTEEDIKEANELVLHSIEINNL 723
>TC83856 weakly similar to PIR|B96656|B96656 unknown protein 41955-40111
[imported] - Arabidopsis thaliana, partial (12%)
Length = 662
Score = 77.8 bits (190), Expect = 1e-14
Identities = 49/161 (30%), Positives = 83/161 (51%), Gaps = 4/161 (2%)
Frame = +2
Query: 331 IIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRTFHAFFRIL-- 388
+IK N+ PD T+N L+ CK+ KI A ++ + M +RG P+I T+ + L
Sbjct: 44 LIK*YLENIKPDVYTFNILVDVFCKSGKISYALKLVDEMHDRGQPPNIVTYSSILDALCK 223
Query: 389 --RVKEEVFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDGIGHDRS 446
RV + V LL K+K+ G P + TY +LI C +L++ + I+ + G
Sbjct: 224 THRVDKAV-ALLTKLKDQGIRPNMHTYTILIDGLCTSGKLEDARNIFEDLLVKGYDITVV 400
Query: 447 SYIVLIHGLFLNCKVEEAYKYYIEMQEKGFLPEPKTESMLQ 487
+YIV+ +G +EA +M+E G +P+ KT +++
Sbjct: 401 TYIVMFYGFCKKGLFDEASALLSKMEENGCIPDAKTYELIK 523
Score = 77.4 bits (189), Expect = 1e-14
Identities = 43/170 (25%), Positives = 85/170 (49%), Gaps = 3/170 (1%)
Frame = +2
Query: 303 NITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEA 362
NI PD +N ++ K+ + A+ L+ +M D P+ +TY+S++ LCK ++D+A
Sbjct: 65 NIKPDVYTFNILVDVFCKSGKISYALKLVDEMHDRGQPPNIVTYSSILDALCKTHRVDKA 244
Query: 363 KEIFNVMLERGISPSIRTFHAFFRILRVK---EEVFELLDKMKELGCNPTIETYIMLIRK 419
+ + ++GI P++ T+ L E+ + + + G + T+ TYI++
Sbjct: 245 VALLTKLKDQGIRPNMHTYTILIDGLCTSGKLEDARNIFEDLLVKGYDITVVTYIVMFYG 424
Query: 420 FCRWRQLDEVKRIWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYI 469
FC+ DE + + M E+G D +Y ++ LF + + A K ++
Sbjct: 425 FCKKGLFDEASALLSKMEENGCIPDAKTYELIKLSLFKKGENDMAEKLHL 574
Score = 58.5 bits (140), Expect = 6e-09
Identities = 37/152 (24%), Positives = 74/152 (48%)
Frame = +2
Query: 230 KNVFPLDTKSFNIILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKL 289
+N+ P D +FNI+++ +C A ++ +EM R ++V+Y+SI+ K+ ++
Sbjct: 62 ENIKP-DVYTFNILVDVFCKS-GKISYALKLVDEMHDRGQPPNIVTYSSILDALCKTHRV 235
Query: 290 YRVLQLFEQMKKRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSL 349
+ + L ++K + I P+ Y +I L + +++A N+ + +TY +
Sbjct: 236 DKAVALLTKLKDQGIRPNMHTYTILIDGLCTSGKLEDARNIFEDLLVKGYDITVVTYIVM 415
Query: 350 IKPLCKARKIDEAKEIFNVMLERGISPSIRTF 381
CK DEA + + M E G P +T+
Sbjct: 416 FYGFCKKGLFDEASALLSKMEENGCIPDAKTY 511
>AW776225 weakly similar to GP|18461169|dbj hypothetical protein~similar to
Arabidopsis thaliana chromosome 5 MCL19.15, partial
(10%)
Length = 396
Score = 76.6 bits (187), Expect = 2e-14
Identities = 43/125 (34%), Positives = 67/125 (53%), Gaps = 3/125 (2%)
Frame = +1
Query: 295 LFEQMKKRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLC 354
+ M R +T + YN VI K +MV +A+ L +M + P+ ITYNSLI LC
Sbjct: 22 ILNTMSHRGVTATVRSYNIVINGFCKIKMVDQAMKLFKEMHHKQIFPNVITYNSLIDGLC 201
Query: 355 KARKIDEAKEIFNVMLERGISPSIRTFHAFFRIL---RVKEEVFELLDKMKELGCNPTIE 411
K+ +I A E+ ++M +RG P I T+ + L + ++ LL K+K+ G P +
Sbjct: 202 KSGRISYALELIDLMHDRGQQPDIITYSSILDALCKNHLVDKAIALLIKLKDQGIRPNMY 381
Query: 412 TYIML 416
TY +L
Sbjct: 382 TYTIL 396
Score = 75.5 bits (184), Expect = 5e-14
Identities = 37/112 (33%), Positives = 69/112 (61%)
Frame = +1
Query: 238 KSFNIILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFE 297
+S+NI++NG+C I A ++++EM ++I +V++Y S+I KS ++ L+L +
Sbjct: 64 RSYNIVINGFCK-IKMVDQAMKLFKEMHHKQIFPNVITYNSLIDGLCKSGRISYALELID 240
Query: 298 QMKKRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSL 349
M R PD Y++++ +L KN +V +A+ L+IK++D + P+ TY L
Sbjct: 241 LMHDRGQQPDIITYSSILDALCKNHLVDKAIALLIKLKDQGIRPNMYTYTIL 396
Score = 73.6 bits (179), Expect = 2e-13
Identities = 40/125 (32%), Positives = 70/125 (56%)
Frame = +1
Query: 257 AERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYNAVIF 316
A+ I MS R + V SY +I+ + K + + ++LF++M + I P+ YN++I
Sbjct: 13 AKSILNTMSHRGVTATVRSYNIVINGFCKIKMVDQAMKLFKEMHHKQIFPNVITYNSLID 192
Query: 317 SLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISP 376
L K+ + A+ LI M D PD ITY+S++ LCK +D+A + + ++GI P
Sbjct: 193 GLCKSGRISYALELIDLMHDRGQQPDIITYSSILDALCKNHLVDKAIALLIKLKDQGIRP 372
Query: 377 SIRTF 381
++ T+
Sbjct: 373 NMYTY 387
Score = 55.1 bits (131), Expect = 7e-08
Identities = 32/128 (25%), Positives = 63/128 (49%), Gaps = 3/128 (2%)
Frame = +1
Query: 327 AVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRTFHAFFR 386
A +++ M VT +YN +I CK + +D+A ++F M + I P++ T+++
Sbjct: 13 AKSILNTMSHRGVTATVRSYNIVINGFCKIKMVDQAMKLFKEMHHKQIFPNVITYNSLID 192
Query: 387 ILRVKEEV---FELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDGIGH 443
L + EL+D M + G P I TY ++ C+ +D+ + +++ GI
Sbjct: 193 GLCKSGRISYALELIDLMHDRGQQPDIITYSSILDALCKNHLVDKAIALLIKLKDQGIRP 372
Query: 444 DRSSYIVL 451
+ +Y +L
Sbjct: 373 NMYTYTIL 396
>AW688886 similar to GP|3258568|gb|A Unknown protein {Arabidopsis thaliana},
partial (11%)
Length = 660
Score = 75.9 bits (185), Expect = 4e-14
Identities = 51/167 (30%), Positives = 91/167 (53%), Gaps = 7/167 (4%)
Frame = +2
Query: 296 FEQMKKRNITPDRKVYN-AVIFSLAKNRMVKEAVNLIIKMEDN--NVTPDAITYNSLIKP 352
F++M + PD YN A++ + +K A NL+ M +++PD +TY +LI+
Sbjct: 11 FKEMTSFDCDPDVVTYNTALLMVCVEXGKIKVAHNLVNGMSKKCKDLSPDVVTYTTLIRG 190
Query: 353 LCKARKIDEAKEIFNVMLERGISPSIRTFHAFFRIL---RVKEEVFELLDKMK-ELGCNP 408
C+ +++DEA +I M RG+ P+I T++ + L + +++ E+L++MK + G P
Sbjct: 191 YCRKQEVDEALDILEEMNGRGLKPNIVTYNTLIKGLCEAQKWDKMKEILEQMKGDGGSIP 370
Query: 409 TIETYIMLIRKFCRWRQLDEVKRIWNAMREDGIGHDRSSYIVLIHGL 455
T+ LI C LDE +++ M++ + D +SY VLI L
Sbjct: 371 DACTFNTLINSHCCAGNLDEAFKVFENMKKLEVSADSASYSVLIRTL 511
Score = 71.2 bits (173), Expect = 9e-13
Identities = 48/169 (28%), Positives = 87/169 (51%), Gaps = 7/169 (4%)
Frame = +2
Query: 261 WEEMSKRRIQHDVVSY-ASIISCYSKSSKLYRVLQLFEQMKKR--NITPDRKVYNAVIFS 317
++EM+ DVV+Y +++ + K+ L M K+ +++PD Y +I
Sbjct: 11 FKEMTSFDCDPDVVTYNTALLMVCVEXGKIKVAHNLVNGMSKKCKDLSPDVVTYTTLIRG 190
Query: 318 LAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVML-ERGISP 376
+ + V EA++++ +M + P+ +TYN+LIK LC+A+K D+ KEI M + G P
Sbjct: 191 YCRKQEVDEALDILEEMNGRGLKPNIVTYNTLIKGLCEAQKWDKMKEILEQMKGDGGSIP 370
Query: 377 SIRTFHAFFR---ILRVKEEVFELLDKMKELGCNPTIETYIMLIRKFCR 422
TF+ +E F++ + MK+L + +Y +LIR C+
Sbjct: 371 DACTFNTLINSHCCAGNLDEAFKVFENMKKLEVSADSASYSVLIRTLCQ 517
Score = 50.8 bits (120), Expect = 1e-06
Identities = 34/132 (25%), Positives = 67/132 (50%), Gaps = 1/132 (0%)
Frame = +2
Query: 236 DTKSFNIILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQL 295
D ++ ++ G+C A I EEM+ R ++ ++V+Y ++I ++ K ++ ++
Sbjct: 158 DVVTYTTLIRGYCRK-QEVDEALDILEEMNGRGLKPNIVTYNTLIKGLCEAQKWDKMKEI 334
Query: 296 FEQMK-KRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLC 354
EQMK PD +N +I S + EA + M+ V+ D+ +Y+ LI+ LC
Sbjct: 335 LEQMKGDGGSIPDACTFNTLINSHCCAGNLDEAFKVFENMKKLEVSADSASYSVLIRTLC 514
Query: 355 KARKIDEAKEIF 366
+ +A+ +F
Sbjct: 515 QKGDYGKAEMLF 550
Score = 38.9 bits (89), Expect = 0.005
Identities = 39/156 (25%), Positives = 67/156 (42%), Gaps = 2/156 (1%)
Frame = +2
Query: 333 KMEDNNVTPDAITYNSLIKPLC-KARKIDEAKEIFNVMLERGISPSIRTFHAFFRILRVK 391
+M + PD +TYN+ + +C + KI A + N M +
Sbjct: 17 EMTSFDCDPDVVTYNTALLMVCVEXGKIKVAHNLVNGMSK-------------------- 136
Query: 392 EEVFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDGIGHDRSSYIVL 451
K K+L +P + TY LIR +CR +++DE I M G+ + +Y L
Sbjct: 137 --------KCKDL--SPDVVTYTTLIRGYCRKQEVDEALDILEEMNGRGLKPNIVTYNTL 286
Query: 452 IHGLFLNCKVEEAYKYYIEMQ-EKGFLPEPKTESML 486
I GL K ++ + +M+ + G +P+ T + L
Sbjct: 287 IKGLCEAQKWDKMKEILEQMKGDGGSIPDACTFNTL 394
Score = 34.7 bits (78), Expect = 0.097
Identities = 28/130 (21%), Positives = 51/130 (38%), Gaps = 3/130 (2%)
Frame = +2
Query: 170 TLLIMIRKYCAVHDVGRAINTFYAFKRFNFQVGLYEFQGLLSALCRYKNVQDAEHLLFCN 229
T +IR YC +V A++ + + + L+ LC + + +L
Sbjct: 167 TYTTLIRGYCRKQEVDEALDILEEMNGRGLKPNIVTYNTLIKGLCEAQKWDKMKEILEQM 346
Query: 230 K---NVFPLDTKSFNIILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKS 286
K P D +FN ++N C + A +++E M K + D SY+ +I +
Sbjct: 347 KGDGGSIP-DACTFNTLINSHC-CAGNLDEAFKVFENMKKLEVSADSASYSVLIRTLCQK 520
Query: 287 SKLYRVLQLF 296
+ LF
Sbjct: 521 GDYGKAEMLF 550
>BF649699 similar to GP|9757911|dbj gb|AAF19552.1~gene_id:MMN10.14~similar to
unknown protein {Arabidopsis thaliana}, partial (11%)
Length = 617
Score = 74.7 bits (182), Expect = 8e-14
Identities = 55/200 (27%), Positives = 91/200 (45%), Gaps = 6/200 (3%)
Frame = +1
Query: 285 KSSKLYRVLQLFEQMKKRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAI 344
K + L ++ + P+ VYNA+I +L K + +A L M N+ + +
Sbjct: 10 KKGNIDSAYDLVVKLGRFGFLPNLFVYNALINALCKGEDLDKAELLYKNMHSMNLPLNDV 189
Query: 345 TYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRTF------HAFFRILRVKEEVFELL 398
TY+ LI CK +D A+ F M+E GI +I + H F L E ++
Sbjct: 190 TYSILIDSFCKRGMLDVAESYFGRMIEDGIRETIYPYNSLINGHCKFGDLSAAEFLY--- 360
Query: 399 DKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDGIGHDRSSYIVLIHGLFLN 458
KM G PT T+ LI +C+ Q+++ +++ M E ++ LI+GL
Sbjct: 361 TKMINEGLEPTATTFTTLISGYCKDLQVEKAFKLYREMNEKEXAPSVYTFTALIYGLCST 540
Query: 459 CKVEEAYKYYIEMQEKGFLP 478
++ EA K + EM E+ P
Sbjct: 541 NEMAEASKLFDEMVERKIKP 600
Score = 67.8 bits (164), Expect = 1e-11
Identities = 48/175 (27%), Positives = 80/175 (45%), Gaps = 6/175 (3%)
Frame = +1
Query: 318 LAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPS 377
L K + A +L++K+ P+ YN+LI LCK +D+A+ ++ M + +
Sbjct: 4 LRKKGNIDSAYDLVVKLGRFGFLPNLFVYNALINALCKGEDLDKAELLYKNMHSMNLPLN 183
Query: 378 IRTFHAFF------RILRVKEEVFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKR 431
T+ +L V E F +M E G TI Y LI C++ L +
Sbjct: 184 DVTYSILIDSFCKRGMLDVAESYF---GRMIEDGIRETIYPYNSLINGHCKFGDLSAAEF 354
Query: 432 IWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFLPEPKTESML 486
++ M +G+ +++ LI G + +VE+A+K Y EM EK P T + L
Sbjct: 355 LYTKMINEGLEPTATTFTTLISGYCKDLQVEKAFKLYREMNEKEXAPSVYTFTAL 519
Score = 63.2 bits (152), Expect = 3e-10
Identities = 59/221 (26%), Positives = 95/221 (42%), Gaps = 3/221 (1%)
Frame = +1
Query: 196 RFNFQVGLYEFQGLLSALCRYKNVQDAEHLLFCNKNVFPLDTKSFNIILNGWCNLIVSAR 255
RF F L+ + L++ALC+ +++ AE LL+ N + S N+ LN
Sbjct: 58 RFGFLPNLFVYNALINALCKGEDLDKAE-LLYKNMH-------SMNLPLND--------- 186
Query: 256 NAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYNAVI 315
V+Y+ +I + K L F +M + I YN++I
Sbjct: 187 ------------------VTYSILIDSFCKRGMLDVAESYFGRMIEDGIRETIYPYNSLI 312
Query: 316 FSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGIS 375
K + A L KM + + P A T+ +LI CK ++++A +++ M E+ +
Sbjct: 313 NGHCKFGDLSAAEFLYTKMINEGLEPTATTFTTLISGYCKDLQVEKAFKLYREMNEKEXA 492
Query: 376 PSIRTFHAFFRILRVKEEVFE---LLDKMKELGCNPTIETY 413
PS+ TF A L E+ E L D+M E PT TY
Sbjct: 493 PSVYTFTALIYGLCSTNEMAEASKLFDEMVERKIKPTXVTY 615
>TC80520 weakly similar to PIR|T02562|T02562 probable salt-inducible protein
[imported] - Arabidopsis thaliana, partial (6%)
Length = 1911
Score = 73.9 bits (180), Expect = 1e-13
Identities = 56/210 (26%), Positives = 98/210 (46%), Gaps = 4/210 (1%)
Frame = +3
Query: 247 WCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITP 306
W + A +++++M R +VVSY+++IS K L LF +M ++ I
Sbjct: 1026 WYCKVRDFAEARKLYDDMCGRGYAENVVSYSTMISGLYLHGKTDEALSLFHEMSRKGIAR 1205
Query: 307 DRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIF 366
D YN++I L + + +A NL+ K+ + P ++ LIK LCK + A +
Sbjct: 1206 DLISYNSLIKGLCQEGELAKATNLLNKLLIQGLEPSVSSFTPLIKCLCKVGDTEGAMRLL 1385
Query: 367 NVMLERGISPSIRTFHAFFRILRVKEEVF----ELLDKMKELGCNPTIETYIMLIRKFCR 422
M +R + P I + H + I K+ F E L KM P ++T+ LI R
Sbjct: 1386 KDMHDRHLEP-IASTHDYMIIGLSKKGDFAQGMEWLLKMLSWKLKPNMQTFEHLIDCLKR 1562
Query: 423 WRQLDEVKRIWNAMREDGIGHDRSSYIVLI 452
+LD++ + + M +G ++S L+
Sbjct: 1563 ENRLDDILIVLDLMFREGYRLEKSRICSLV 1652
Score = 68.6 bits (166), Expect = 6e-12
Identities = 52/223 (23%), Positives = 99/223 (44%), Gaps = 8/223 (3%)
Frame = +1
Query: 241 NIILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYR---VLQLFE 297
N L G+ L+ E +++ + + S +S C K+ R V +L+E
Sbjct: 364 NDSLEGYVRLLGENGMVEEVFDVFVSLKKVGFLPSASSFNVCLLACLKVGRTDLVWKLYE 543
Query: 298 QMKKRNI--TPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCK 355
M + + D + +I + V L+ ++ + + D +N+LI CK
Sbjct: 544 LMIESGVGVNIDVETVGCLIKAFCAENKVFNGYELLRQVLEKGLCVDNTVFNALINGFCK 723
Query: 356 ARKIDEAKEIFNVMLERGISPSIRTFHAFFRIL---RVKEEVFELLDKMKELGCNPTIET 412
++ D EI ++M+ +PSI T+ L R +E F + + +K+ G P
Sbjct: 724 QKQYDRVSEILHIMIAMKCNPSIYTYQEIINGLLKRRKNDEAFRVFNDLKDRGYFPDRVM 903
Query: 413 YIMLIRKFCRWRQLDEVKRIWNAMREDGIGHDRSSYIVLIHGL 455
Y +I+ FC L E +++W M + G+ + +Y V+I+G+
Sbjct: 904 YTTVIKGFCDMGLLAEARKLWFEMIQKGLVPNEYTYNVMIYGI 1032
Score = 38.9 bits (89), Expect = 0.005
Identities = 18/56 (32%), Positives = 31/56 (55%)
Frame = +3
Query: 420 FCRWRQLDEVKRIWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKG 475
+C+ R E +++++ M G + SY +I GL+L+ K +EA + EM KG
Sbjct: 1029 YCKVRDFAEARKLYDDMCGRGYAENVVSYSTMISGLYLHGKTDEALSLFHEMSRKG 1196
>BE248944 similar to GP|11994509|dbj gb|AAF27119.1~gene_id:F4B12.11~similar
to unknown protein {Arabidopsis thaliana}, partial (8%)
Length = 357
Score = 72.4 bits (176), Expect = 4e-13
Identities = 36/114 (31%), Positives = 66/114 (57%), Gaps = 2/114 (1%)
Frame = +2
Query: 334 MEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERG--ISPSIRTFHAFFRILRVK 391
M++ + P+ TYNSLIK LCK R++++ E+ M R P+ T+ + L+
Sbjct: 14 MKERDCLPNVATYNSLIKHLCKIRRMEKVYELVEDMERRSGDCLPNGVTYSYLLQSLKAP 193
Query: 392 EEVFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDGIGHDR 445
EEV +L++M+ GC + + +++R + +W LD +++ W+ M +G+G DR
Sbjct: 194 EEVPAVLERMERNGCAMSDDICNLVLRLYMKWDDLDGLRKTWDEMERNGLGPDR 355
>AW695801 weakly similar to PIR|T01622|T01 probable salt-inducible protein
At2g18940 [imported] - Arabidopsis thaliana, partial
(18%)
Length = 655
Score = 70.1 bits (170), Expect = 2e-12
Identities = 47/194 (24%), Positives = 93/194 (47%), Gaps = 6/194 (3%)
Frame = +3
Query: 217 KNVQDAEHLLFCNKNVFPLDTKSFNIILNGWCNLIVSARNAERIWEEMSKRRI---QHDV 273
K ++ A H L N + LD +++N ++ V + E+ E + + Q ++
Sbjct: 9 KGMERAFHQL--QNNGYKLDM----VVINSMLSMFVRNQKLEKAHEMLDVIHVSGLQPNL 170
Query: 274 VSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIK 333
V+Y S+I Y++ ++ ++ + ++ I+PD YN VI K +V+EA+ ++ +
Sbjct: 171 VTYNSLIDLYARVGDCWKAEEMLKDIQNSGISPDVVSYNTVIKGFCKKGLVQEAIRILSE 350
Query: 334 MEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRTFHAF---FRILRV 390
M N V P IT+N+ + EA E+ M+E G P+ T+ + +
Sbjct: 351 MTANGVQPCPITFNTFMSCYAGNGLFAEADEVIRYMIEHGCMPNELTYKIVIDGYIKAKK 530
Query: 391 KEEVFELLDKMKEL 404
+E + + K+KE+
Sbjct: 531 HKEAMDFVSKIKEI 572
Score = 52.8 bits (125), Expect = 3e-07
Identities = 42/184 (22%), Positives = 85/184 (45%), Gaps = 6/184 (3%)
Frame = +3
Query: 296 FEQMKKRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCK 355
F Q++ D V N+++ +N+ +++A ++ + + + P+ +TYNSLI +
Sbjct: 27 FHQLQNNGYKLDMVVINSMLSMFVRNQKLEKAHEMLDVIHVSGLQPNLVTYNSLIDLYAR 206
Query: 356 ARKIDEAKEIFNVMLERGISPSIRTFHAFFRILRVK---EEVFELLDKMKELGCNP---T 409
+A+E+ + GISP + +++ + K +E +L +M G P T
Sbjct: 207 VGDCWKAEEMLKDIQNSGISPDVVSYNTVIKGFCKKGLVQEAIRILSEMTANGVQPCPIT 386
Query: 410 IETYIMLIRKFCRWRQLDEVKRIWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYI 469
T++ + + DEV R M E G + +Y ++I G K +EA +
Sbjct: 387 FNTFMSCYAGNGLFAEADEVIRY---MIEHGCMPNELTYKIVIDGYIKAKKHKEAMDFVS 557
Query: 470 EMQE 473
+++E
Sbjct: 558 KIKE 569
>TC83956 weakly similar to GP|15982931|gb|AAL09812.1 AT3g53700/F4P12_400
{Arabidopsis thaliana}, partial (18%)
Length = 762
Score = 67.8 bits (164), Expect = 1e-11
Identities = 44/187 (23%), Positives = 90/187 (47%), Gaps = 3/187 (1%)
Frame = +1
Query: 272 DVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYNAVIFSLAKNRMVKEAVNLI 331
+ ++A++I ++ ++ +L++ E + PD YN + +L ++ +K L
Sbjct: 190 NATTFATLIQSFTNFHEIENLLKILEN--ELGFKPDTNFYNIALNALVEDNKLKLVEMLH 363
Query: 332 IKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRTFHAFFRILRVK 391
KM + + D T+N LIK LCKA ++ A + M G+ P TF + +
Sbjct: 364 SKMVNEGIVLDVSTFNVLIKALCKAHQLRPAILMLEEMANHGLKPDEITFTTLMQGFIEE 543
Query: 392 EEVFELLDKMKEL---GCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDGIGHDRSSY 448
++ L K++ GC T + +L+ FC+ +++E R + E+G D+ ++
Sbjct: 544 GDLNGALKMKKQMLGYGCLLTNVSVKVLVNGFCKEGRVEEALRFVLEVSEEGFSPDQGTF 723
Query: 449 IVLIHGL 455
L++G+
Sbjct: 724 NSLVNGV 744
Score = 58.2 bits (139), Expect = 8e-09
Identities = 46/233 (19%), Positives = 101/233 (42%), Gaps = 2/233 (0%)
Frame = +1
Query: 128 HSVREYHSMISVLGKMRRFDTAWALVEEMRRGKTGESIVTPQTLLIMIRKYCAVHDVGRA 187
H + +++ L + FD+ L++++ K+ SI T +I+ + H++
Sbjct: 82 HPLPPNETLLLQLTQSSSFDSITTLLKQL---KSSGSIPNATTFATLIQSFTNFHEIENL 252
Query: 188 INTFYAFKRFNFQVGLYEFQGLLSALCRYKNVQDAE--HLLFCNKNVFPLDTKSFNIILN 245
+ F Y L+AL ++ E H N+ + LD +FN+++
Sbjct: 253 LKILENELGFKPDTNFYNIA--LNALVEDNKLKLVEMLHSKMVNEGIV-LDVSTFNVLIK 423
Query: 246 GWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNIT 305
C R A + EEM+ ++ D +++ +++ + + L L++ +QM
Sbjct: 424 ALCKAH-QLRPAILMLEEMANHGLKPDEITFTTLMQGFIEEGDLNGALKMKKQMLGYGCL 600
Query: 306 PDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARK 358
++ K V+EA+ ++++ + +PD T+NSL+ +C+ K
Sbjct: 601 LTNVSVKVLVNGFCKEGRVEEALRFVLEVSEEGFSPDQGTFNSLVNGVCRIWK 759
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.137 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,059,400
Number of Sequences: 36976
Number of extensions: 234827
Number of successful extensions: 1906
Number of sequences better than 10.0: 133
Number of HSP's better than 10.0 without gapping: 1453
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1727
length of query: 550
length of database: 9,014,727
effective HSP length: 101
effective length of query: 449
effective length of database: 5,280,151
effective search space: 2370787799
effective search space used: 2370787799
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC123593.8


