

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123593.7 + phase: 0
(467 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
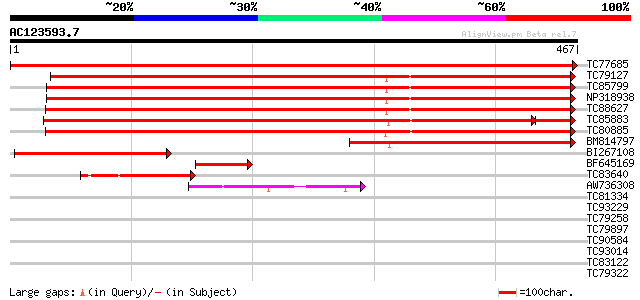
Score E
Sequences producing significant alignments: (bits) Value
TC77685 apyrase-like protein [Medicago truncatula] 928 0.0
TC79127 apyrase-like protein [Medicago truncatula] 554 e-158
TC85799 apyrase-like protein [Medicago truncatula] 538 e-153
NP318938 NP318938|AF288132.1|AAK15160.1 putative apyrase [Medica... 533 e-152
TC88627 apyrase-like protein [Medicago truncatula] 525 e-149
TC85883 apyrase-like protein [Medicago truncatula] 494 e-149
TC80885 apyrase-like protein [Medicago truncatula] 516 e-147
BM814797 similar to GP|10717159|gb apyrase 2 {Pisum sativum}, pa... 312 2e-85
BI267108 homologue to GP|10717159|gb apyrase 2 {Pisum sativum}, ... 169 2e-42
BF645169 similar to GP|10717159|gb| apyrase 2 {Pisum sativum}, p... 65 7e-11
TC83640 similar to GP|14423550|gb|AAK62457.1 putative nucleoside... 62 5e-10
AW736308 similar to GP|19920124|gb| Putative nucleoside phosphat... 61 8e-10
TC81334 weakly similar to GP|19423992|gb|AAL87274.1 putative ABC... 34 0.14
TC93229 similar to GP|14517424|gb|AAK62602.1 AT5g41370/MYC6_8 {A... 32 0.52
TC79258 similar to GP|21592967|gb|AAM64916.1 putative GDSL-motif... 32 0.68
TC79897 similar to PIR|E84603|E84603 probable protein transport ... 30 1.5
TC90584 similar to GP|22136810|gb|AAM91749.1 unknown protein {Ar... 30 2.0
TC93014 similar to PIR|T05880|T05880 hypothetical protein T29A15... 30 2.0
TC83122 similar to PIR|C84658|C84658 3-beta-hydroxysteroid dehyd... 30 2.6
TC79322 similar to GP|13605815|gb|AAK32893.1 At2g32700/F24L7.16 ... 29 4.4
>TC77685 apyrase-like protein [Medicago truncatula]
Length = 2172
Score = 928 bits (2399), Expect = 0.0
Identities = 467/467 (100%), Positives = 467/467 (100%)
Frame = +1
Query: 1 MLKRPTRQESLSDKIYRFRGTLLVVSIPILLITFVLYMMPSSSNYDSAGDYALLNRKLSP 60
MLKRPTRQESLSDKIYRFRGTLLVVSIPILLITFVLYMMPSSSNYDSAGDYALLNRKLSP
Sbjct: 346 MLKRPTRQESLSDKIYRFRGTLLVVSIPILLITFVLYMMPSSSNYDSAGDYALLNRKLSP 525
Query: 61 DKKSGGSYAVIFDAGSSGSRVHVFHFDQNLDLVHIGKDLELFEQLKPGLSAYAQKPQQAA 120
DKKSGGSYAVIFDAGSSGSRVHVFHFDQNLDLVHIGKDLELFEQLKPGLSAYAQKPQQAA
Sbjct: 526 DKKSGGSYAVIFDAGSSGSRVHVFHFDQNLDLVHIGKDLELFEQLKPGLSAYAQKPQQAA 705
Query: 121 ESLVSLLEKAEGVVPRELRSKTPVRIGATAGLRALEGDASDKILRAVRDLLKHRSSFKSD 180
ESLVSLLEKAEGVVPRELRSKTPVRIGATAGLRALEGDASDKILRAVRDLLKHRSSFKSD
Sbjct: 706 ESLVSLLEKAEGVVPRELRSKTPVRIGATAGLRALEGDASDKILRAVRDLLKHRSSFKSD 885
Query: 181 ADAVTVLDGTQEGAYQWVTINYLLGNLGKDYSKTVGVVDLGGGSVQMAYAISESEAAMAP 240
ADAVTVLDGTQEGAYQWVTINYLLGNLGKDYSKTVGVVDLGGGSVQMAYAISESEAAMAP
Sbjct: 886 ADAVTVLDGTQEGAYQWVTINYLLGNLGKDYSKTVGVVDLGGGSVQMAYAISESEAAMAP 1065
Query: 241 QVMDGEDPYVKEMFLRGRKYYLYVHSYLRYGLLAARAEILKVAGDAENPCILSGSDGTYK 300
QVMDGEDPYVKEMFLRGRKYYLYVHSYLRYGLLAARAEILKVAGDAENPCILSGSDGTYK
Sbjct: 1066QVMDGEDPYVKEMFLRGRKYYLYVHSYLRYGLLAARAEILKVAGDAENPCILSGSDGTYK 1245
Query: 301 YGGKSFKASSSGASLNECKSVAHKALKVNESTCTHMKCTFGGIWNGGGGDGQKNLFVASF 360
YGGKSFKASSSGASLNECKSVAHKALKVNESTCTHMKCTFGGIWNGGGGDGQKNLFVASF
Sbjct: 1246YGGKSFKASSSGASLNECKSVAHKALKVNESTCTHMKCTFGGIWNGGGGDGQKNLFVASF 1425
Query: 361 FFDRAAEAGFVDPNSPVAIVRPADFEDAAKQACQTKLENAKSTYPRVEEGNLPYLCMDLV 420
FFDRAAEAGFVDPNSPVAIVRPADFEDAAKQACQTKLENAKSTYPRVEEGNLPYLCMDLV
Sbjct: 1426FFDRAAEAGFVDPNSPVAIVRPADFEDAAKQACQTKLENAKSTYPRVEEGNLPYLCMDLV 1605
Query: 421 YQYTLLVDGFGIYPWQEITLVKKVKYDDALVEAAWPLGSAIEAVSST 467
YQYTLLVDGFGIYPWQEITLVKKVKYDDALVEAAWPLGSAIEAVSST
Sbjct: 1606YQYTLLVDGFGIYPWQEITLVKKVKYDDALVEAAWPLGSAIEAVSST 1746
>TC79127 apyrase-like protein [Medicago truncatula]
Length = 1501
Score = 554 bits (1427), Expect = e-158
Identities = 269/437 (61%), Positives = 346/437 (78%), Gaps = 4/437 (0%)
Frame = +3
Query: 34 FVLYMMPSSSNYDSAGDYALL-NRKLSPDKKSGGSYAVIFDAGSSGSRVHVFHFDQNLDL 92
F L++MPS S + G+ LL NRK+ P +++ SYAV+FDAGS+GSRVHV+HFDQNL+L
Sbjct: 81 FYLFIMPSISYSQNLGNNILLTNRKIFPKQETISSYAVVFDAGSTGSRVHVYHFDQNLNL 260
Query: 93 VHIGKDLELFEQLKPGLSAYAQKPQQAAESLVSLLEKAEGVVPRELRSKTPVRIGATAGL 152
+H+GKD+E + + PGLSAYA P+QAA+SL+ LLE+AE VVP + RSKTPVR+GATAGL
Sbjct: 261 LHVGKDVEFYNKTTPGLSAYADNPEQAAKSLIPLLEQAESVVPEDQRSKTPVRLGATAGL 440
Query: 153 RALEGDASDKILRAVRDLLKHRSSFKSDADAVTVLDGTQEGAYQWVTINYLLGNLGKDYS 212
R L GDAS+KIL++VRDLL +RS+F DAV+++DGTQEG+Y WVT+NY LG LGK ++
Sbjct: 441 RLLNGDASEKILQSVRDLLSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYALGTLGKKFT 620
Query: 213 KTVGVVDLGGGSVQMAYAISESEAAMAPQVMDGEDPYVKEMFLRGRKYYLYVHSYLRYGL 272
KTVGV+DLGGGSVQMAYA+S + A AP+V DG+DPY+K++ L+G+KY LYVHSYL +G
Sbjct: 621 KTVGVMDLGGGSVQMAYAVSRNTAKNAPKVADGDDPYIKKLVLKGKKYDLYVHSYLHFGT 800
Query: 273 LAARAEILKVAGDAENPCILSGSDGTYKYGGKSFKAS--SSGASLNECKSVAHKALKVNE 330
A+RAEILKV ++ NPCIL+G DGTY+Y G+ FKA+ +SGAS +CK + H+ALK+N
Sbjct: 801 EASRAEILKVTHNSPNPCILAGFDGTYRYAGEEFKANALASGASFKKCKKIVHQALKLN- 977
Query: 331 STCTHMKCTFGGIWNGGGGDGQKNLFVASFFFDRAAEAGFVDPNSPVAIVRPADFEDAAK 390
C + CTFGGIWNGGGG GQ+ LF ASFFF AAE G VDPN P +RP DFE AK
Sbjct: 978 YPCPYQNCTFGGIWNGGGGSGQRKLFAASFFFYLAAEVGMVDPNKPNFKIRPVDFESEAK 1157
Query: 391 QACQTKLENAKSTYPRVEEGNL-PYLCMDLVYQYTLLVDGFGIYPWQEITLVKKVKYDDA 449
+AC E+AKS+YP + + N+ Y+CMDL+YQY LLVDGFG+ P QEIT K+++Y D+
Sbjct: 1158KACALNFEDAKSSYPFLAKKNIASYVCMDLIYQYVLLVDGFGLDPLQEITAGKQIEYQDS 1337
Query: 450 LVEAAWPLGSAIEAVSS 466
LVEAAWPLG+A+EA+SS
Sbjct: 1338LVEAAWPLGNAVEAISS 1388
>TC85799 apyrase-like protein [Medicago truncatula]
Length = 1674
Score = 538 bits (1387), Expect = e-153
Identities = 265/441 (60%), Positives = 345/441 (78%), Gaps = 5/441 (1%)
Frame = +3
Query: 31 LITFVLYMMPSSSNYDSAGDYALL-NRKLSPDKKSGGSYAVIFDAGSSGSRVHVFHFDQN 89
LITF+L++MPS S G+ LL NRK+ P ++ SYAV+FDAGS+GSRVHV+HFDQN
Sbjct: 222 LITFLLFIMPSISYSQYLGNNILLTNRKIFPKQEPISSYAVVFDAGSTGSRVHVYHFDQN 401
Query: 90 LDLVHIGKDLELFEQLKPGLSAYAQKPQQAAESLVSLLEKAEGVVPRELRSKTPVRIGAT 149
L+L+H+GKD+E + + PGLSAYA P++AA+SL+ LLE+AE VVP + RSKTP+R+GAT
Sbjct: 402 LNLLHVGKDVEFYNKTTPGLSAYADNPEEAAKSLIPLLEQAESVVPEDQRSKTPIRLGAT 581
Query: 150 AGLRALEGDASDKILRAVRDLLKHRSSFKSDADAVTVLDGTQEGAYQWVTINYLLGNLGK 209
AGLR L GDAS+KIL++VRDL +RS+F DAV+++DGTQEG+Y WVT+NY LG LGK
Sbjct: 582 AGLRLLNGDASEKILQSVRDLFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYALGKLGK 761
Query: 210 DYSKTVGVVDLGGGSVQMAYAISESEAAMAPQVMDGEDPYVKEMFLRGRKYYLYVHSYLR 269
++KTVGV+DLGGGSVQMAYA+S+ A AP+V DGEDPY+K++ L+G+KY LYVHSYL
Sbjct: 762 KFTKTVGVMDLGGGSVQMAYAVSKYTAKNAPKVADGEDPYIKKLVLKGKKYDLYVHSYLH 941
Query: 270 YGLLAARAEILKVAGDAENPCILSGSDGTYKYGGKSFKAS--SSGASLNECKSVAHKALK 327
+G A+RAEILKV ++ NPCIL+G DGTY Y G+ FKA+ +SGA+ +CK + +ALK
Sbjct: 942 FGREASRAEILKVTHNSPNPCILAGFDGTYTYAGEEFKANAPASGANFKKCKKIVREALK 1121
Query: 328 VNESTCTHMKCTFGGIWNGGGGDGQKNLFVASFFFDRAAEAGFVDPNSPVAI-VRPADFE 386
+N C + CTFGGIW+GGGG GQ+ LF AS FF A + G VDPN+P ++ +RP D E
Sbjct: 1122LN-YPCPYQNCTFGGIWSGGGGSGQRILFAASSFFYLAEDIGLVDPNTPYSLTLRPVDLE 1298
Query: 387 DAAKQACQTKLENAKSTYPRVEEGNL-PYLCMDLVYQYTLLVDGFGIYPWQEITLVKKVK 445
AK+AC LE AKSTYP + + N+ Y+CMDL+YQY LLVDGFG+ P QEIT K+++
Sbjct: 1299TEAKKACTLNLEEAKSTYPLLVDFNIVEYVCMDLIYQYVLLVDGFGLDPLQEITAGKQIE 1478
Query: 446 YDDALVEAAWPLGSAIEAVSS 466
Y D+LVEAAWPLG+A+EA+SS
Sbjct: 1479YQDSLVEAAWPLGNAVEAISS 1541
>NP318938 NP318938|AF288132.1|AAK15160.1 putative apyrase [Medicago
truncatula]
Length = 1401
Score = 533 bits (1374), Expect = e-152
Identities = 260/440 (59%), Positives = 339/440 (76%), Gaps = 4/440 (0%)
Frame = +1
Query: 31 LITFVLYMMPSSSNYDSAGDYALL-NRKLSPDKKSGGSYAVIFDAGSSGSRVHVFHFDQN 89
LITF+L++MPS S G+ LL NRK+ P ++ SYAV+FDAGS+GSRVHV+HFDQN
Sbjct: 49 LITFLLFIMPSISYSQYLGNNILLTNRKIFPKQEPISSYAVVFDAGSTGSRVHVYHFDQN 228
Query: 90 LDLVHIGKDLELFEQLKPGLSAYAQKPQQAAESLVSLLEKAEGVVPRELRSKTPVRIGAT 149
L+L+H+GKD+E + + PGLSAYA P++AA+SL+ LLE+AE VVP + RSKTP+R+GAT
Sbjct: 229 LNLLHVGKDVEFYNKTTPGLSAYADNPEEAAKSLIPLLEQAESVVPEDQRSKTPIRLGAT 408
Query: 150 AGLRALEGDASDKILRAVRDLLKHRSSFKSDADAVTVLDGTQEGAYQWVTINYLLGNLGK 209
AGLR L GDAS+KIL++VRDL +RS+F DAV+++DGTQEG Y WVT+NY LGNLGK
Sbjct: 409 AGLRLLNGDASEKILQSVRDLFSNRSTFNVQPDAVSIIDGTQEGCYLWVTVNYALGNLGK 588
Query: 210 DYSKTVGVVDLGGGSVQMAYAISESEAAMAPQVMDGEDPYVKEMFLRGRKYYLYVHSYLR 269
++KTVGV+D+GGGSVQMAYA+S+ A AP+V DGEDPY+K++ L+G+ Y LYVHSYL
Sbjct: 589 KFTKTVGVMDVGGGSVQMAYAVSKYTAKNAPKVADGEDPYIKKLVLKGKPYDLYVHSYLH 768
Query: 270 YGLLAARAEILKVAGDAENPCILSGSDGTYKYGGKSFKAS--SSGASLNECKSVAHKALK 327
+G A+RAEI+KV + NPC+L+G DGTY Y G+ FKA +SGA+ N CK + KALK
Sbjct: 769 FGREASRAEIMKVTRSSPNPCLLAGFDGTYTYAGEEFKAKAPASGANFNGCKKIIRKALK 948
Query: 328 VNESTCTHMKCTFGGIWNGGGGDGQKNLFVASFFFDRAAEAGFVDPNSPVAIVRPADFED 387
+N C + CTFGGIWNGGGG+GQK+LF +S FF + G VDP +P +RP D
Sbjct: 949 LN-YPCPYQNCTFGGIWNGGGGNGQKHLFASSSFFYLPEDVGMVDPKTPNFKIRPVDLVS 1125
Query: 388 AAKQACQTKLENAKSTYPRVEEGNL-PYLCMDLVYQYTLLVDGFGIYPWQEITLVKKVKY 446
AK+AC E+AKSTYP + + N+ Y+CMDL+YQY LLVDGFG+ P QEIT K+++Y
Sbjct: 1126EAKKACALNFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDGFGLDPLQEITSGKEIEY 1305
Query: 447 DDALVEAAWPLGSAIEAVSS 466
DA++EAAWPLG+A+EA+SS
Sbjct: 1306QDAVLEAAWPLGNAVEAISS 1365
>TC88627 apyrase-like protein [Medicago truncatula]
Length = 1614
Score = 525 bits (1351), Expect = e-149
Identities = 254/440 (57%), Positives = 335/440 (75%), Gaps = 3/440 (0%)
Frame = +1
Query: 30 LLITFVLYMMPSSSNYDSAGDYALLNRKLSPDKKSGGSYAVIFDAGSSGSRVHVFHFDQN 89
L+ T +L +MP+ ++ G+ L NRK+ +++ SYAV+FDAGS+GSR+HV+HFDQN
Sbjct: 91 LITTVLLLLMPAITSSQYLGNNLLTNRKIFQKQETISSYAVVFDAGSTGSRIHVYHFDQN 270
Query: 90 LDLVHIGKDLELFEQLKPGLSAYAQKPQQAAESLVSLLEKAEGVVPRELRSKTPVRIGAT 149
LDL+HIGKD+E F ++ PGLS+YA P+QAA+SL+ LL++AE VVP +L KTP+R+GAT
Sbjct: 271 LDLLHIGKDVEFFNKITPGLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHKTPIRLGAT 450
Query: 150 AGLRALEGDASDKILRAVRDLLKHRSSFKSDADAVTVLDGTQEGAYQWVTINYLLGNLGK 209
AGLR L GDAS+KIL+AVRD+ +RS+F DAV+++DGTQEG+Y WVT+NY LGNLGK
Sbjct: 451 AGLRLLNGDASEKILQAVRDMFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYALGNLGK 630
Query: 210 DYSKTVGVVDLGGGSVQMAYAISESEAAMAPQVMDGEDPYVKEMFLRGRKYYLYVHSYLR 269
Y+KTVGV+DLGGGSVQMAYA+S+ A AP+V DG DPY+K++ L+G+ Y LYVHSYL
Sbjct: 631 KYTKTVGVMDLGGGSVQMAYAVSKKTAKNAPKVADGVDPYIKKLVLKGKPYDLYVHSYLH 810
Query: 270 YGLLAARAEILKVAGDAENPCILSGSDGTYKYGGKSFKAS--SSGASLNECKSVAHKALK 327
+G A+RAEI+KV + N CIL+G DGTY Y G+ FKA +SGA+ N CK + KALK
Sbjct: 811 FGREASRAEIMKVTRSSPNTCILTGFDGTYTYAGEEFKAKAPASGANFNGCKKIIRKALK 990
Query: 328 VNESTCTHMKCTFGGIWNGGGGDGQKNLFVASFFFDRAAEAGFVDPNSPVAIVRPADFED 387
+N C + CTFGGIWNGGGG+GQK+L +S FF + G VDP +P +RP D
Sbjct: 991 LN-YPCPYQNCTFGGIWNGGGGNGQKHLLASSSFFYLPEDVGMVDPKTPNFKIRPVDLVS 1167
Query: 388 AAKQACQTKLENAKSTYPRVEEGNL-PYLCMDLVYQYTLLVDGFGIYPWQEITLVKKVKY 446
AK+AC E+AKSTYP + + N+ Y+CMDL+YQY LLVDGFG+ P QEIT K+++Y
Sbjct: 1168EAKKACALNFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDGFGLDPLQEITSGKEIEY 1347
Query: 447 DDALVEAAWPLGSAIEAVSS 466
DA++EAAWPLG+A+EA+SS
Sbjct: 1348QDAVLEAAWPLGNAVEAISS 1407
>TC85883 apyrase-like protein [Medicago truncatula]
Length = 1848
Score = 494 bits (1272), Expect(2) = e-149
Identities = 240/408 (58%), Positives = 311/408 (75%), Gaps = 2/408 (0%)
Frame = +3
Query: 29 ILLITFVLYMMPSSSNYDSAGDYALLNRKLSPDKKSGGSYAVIFDAGSSGSRVHVFHFDQ 88
I LITF+L++MP+ S+ G+ L NRK+ P +++ SYAV+FDAGS+GSRVHV+HFDQ
Sbjct: 258 IKLITFLLFLMPTISSSQYLGNNILTNRKIFPKQETLTSYAVVFDAGSTGSRVHVYHFDQ 437
Query: 89 NLDLVHIGKDLELFEQLKPGLSAYAQKPQQAAESLVSLLEKAEGVVPRELRSKTPVRIGA 148
NLDL+HIG D+E + + PGLSAYA P++AAESL+ LLE+AE VVP L+ KTPV++GA
Sbjct: 438 NLDLLHIGNDVEFYNKTTPGLSAYADNPKEAAESLIPLLEQAERVVPVNLQPKTPVKLGA 617
Query: 149 TAGLRALEGDASDKILRAVRDLLKHRSSFKSDADAVTVLDGTQEGAYQWVTINYLLGNLG 208
TAGLR L+G++S+ IL AV LLK RS+F +DAV ++DGTQEG+Y WVTINY+LGNLG
Sbjct: 618 TAGLRLLDGNSSELILEAVSSLLKKRSTFNVQSDAVGIIDGTQEGSYLWVTINYVLGNLG 797
Query: 209 KDYSKTVGVVDLGGGSVQMAYAISESEAAMAPQVMDGEDPYVKEMFLRGRKYYLYVHSYL 268
KD+S+TV V DLGGGSVQM YA+S +A APQV GEDPY+K++ L+G+KYYLYVHSYL
Sbjct: 798 KDFSETVAVADLGGGSVQMVYAVSREQAKKAPQVPQGEDPYIKKIVLKGKKYYLYVHSYL 977
Query: 269 RYGLLAARAEILKVAGDAENPCILSGSDGTYKYGGKSFKASS--SGASLNECKSVAHKAL 326
R+G A+RAEILKV + NPCIL+G GTY Y G+ +KA S SG++ +ECK + KAL
Sbjct: 978 RFGKEASRAEILKVTNGSPNPCILAGYHGTYTYSGEEYKAFSPASGSNFDECKEIILKAL 1157
Query: 327 KVNESTCTHMKCTFGGIWNGGGGDGQKNLFVASFFFDRAAEAGFVDPNSPVAIVRPADFE 386
KVN+ C + KC+FGGIWNGGGG GQK L+V S F+ DPN P + +R D +
Sbjct: 1158KVND-PCPYGKCSFGGIWNGGGGSGQKTLYVTSSFYYVPTGVNIADPNKPNSKIRIEDLK 1334
Query: 387 DAAKQACQTKLENAKSTYPRVEEGNLPYLCMDLVYQYTLLVDGFGIYP 434
A+Q C+TK ++AK+TYP + E +LPY C+DL+YQYTL VDGFG+ P
Sbjct: 1335TGAEQVCKTKYKDAKATYPLIYEDSLPYACLDLIYQYTLFVDGFGLDP 1478
Score = 51.6 bits (122), Expect(2) = e-149
Identities = 22/33 (66%), Positives = 29/33 (87%)
Frame = +1
Query: 434 PWQEITLVKKVKYDDALVEAAWPLGSAIEAVSS 466
P QEIT+ +++Y DALV+AAWPLG+AIEA+SS
Sbjct: 1477 PLQEITVANQIEYQDALVDAAWPLGNAIEAISS 1575
>TC80885 apyrase-like protein [Medicago truncatula]
Length = 1451
Score = 516 bits (1329), Expect = e-147
Identities = 243/439 (55%), Positives = 332/439 (75%), Gaps = 2/439 (0%)
Frame = +2
Query: 30 LLITFVLYMMPSSSNYDSAGDYALLNRKLSPDKKSGGSYAVIFDAGSSGSRVHVFHFDQN 89
L+ TF+L +MP+ ++ G+ L NRK+ +++ SYAVIFDAGS+G+RVHV+HFDQN
Sbjct: 23 LITTFLLLLMPAITSSQYLGNNLLTNRKIFQKQETLTSYAVIFDAGSTGTRVHVYHFDQN 202
Query: 90 LDLVHIGKDLELFEQLKPGLSAYAQKPQQAAESLVSLLEKAEGVVPRELRSKTPVRIGAT 149
LDL+HIG D+E +++KPGLSAYA P+QAA+SL+ LLE+AE V+P ++ KTP+R+GAT
Sbjct: 203 LDLLHIGNDIEFVDKIKPGLSAYADNPEQAAKSLLPLLEEAEDVIPEDMHPKTPLRLGAT 382
Query: 150 AGLRALEGDASDKILRAVRDLLKHRSSFKSDADAVTVLDGTQEGAYQWVTINYLLGNLGK 209
AGLR L GDA++KIL+A R++ +RS+ +DAV+++DGTQEG+Y WVT+NY+LGNLGK
Sbjct: 383 AGLRLLNGDAAEKILQATRNMFSNRSTLNVQSDAVSIIDGTQEGSYMWVTVNYILGNLGK 562
Query: 210 DYSKTVGVVDLGGGSVQMAYAISESEAAMAPQVMDGEDPYVKEMFLRGRKYYLYVHSYLR 269
++KTVGV+DLGGGSVQM YA+S+ A AP+V DGEDPY+K++ L+G++Y LYVHSYLR
Sbjct: 563 SFTKTVGVIDLGGGSVQMTYAVSKKTAKNAPKVADGEDPYIKKLVLKGKQYDLYVHSYLR 742
Query: 270 YGLLAARAEILKVAGDAENPCILSGSDGTYKYGGKSFKA--SSSGASLNECKSVAHKALK 327
+G A RA++L + NPCIL G +GT+ Y G +KA SSG++ NECK + K LK
Sbjct: 743 FGKEATRAQVLNATNGSANPCILPGFNGTFTYSGVEYKAFSPSSGSNFNECKEIILKVLK 922
Query: 328 VNESTCTHMKCTFGGIWNGGGGDGQKNLFVASFFFDRAAEAGFVDPNSPVAIVRPADFED 387
VN+ C + CTF GIWNGGGG GQK LFV S F + G V+PN P + + P DFE
Sbjct: 923 VND-PCPYSSCTFSGIWNGGGGSGQKKLFVTSAFAYLTEDVGMVEPNKPNSTLHPIDFEI 1099
Query: 388 AAKQACQTKLENAKSTYPRVEEGNLPYLCMDLVYQYTLLVDGFGIYPWQEITLVKKVKYD 447
AK+AC E+ KSTYPR+ E PY+CMDL+YQ+ LLV GFG+ P +EIT+ + ++Y
Sbjct: 1100EAKRACALNFEDVKSTYPRLTEAKRPYVCMDLLYQHVLLVHGFGLSPRKEITVGEGIQYQ 1279
Query: 448 DALVEAAWPLGSAIEAVSS 466
+++VEAAWPLG+A+EA+S+
Sbjct: 1280NSVVEAAWPLGTAVEAIST 1336
>BM814797 similar to GP|10717159|gb apyrase 2 {Pisum sativum}, partial (42%)
Length = 690
Score = 312 bits (800), Expect = 2e-85
Identities = 152/188 (80%), Positives = 168/188 (88%), Gaps = 2/188 (1%)
Frame = +1
Query: 281 KVAGDAENPCILSGSDGTYKYGGKSFKASSS--GASLNECKSVAHKALKVNESTCTHMKC 338
K + D NPCIL+G DG+YKYGGKSF ASSS G+SLNECKS+A AL VNESTC HMKC
Sbjct: 4 KASDDFGNPCILAGYDGSYKYGGKSFNASSSPSGSSLNECKSIALNALTVNESTCAHMKC 183
Query: 339 TFGGIWNGGGGDGQKNLFVASFFFDRAAEAGFVDPNSPVAIVRPADFEDAAKQACQTKLE 398
TFGGIWNGGGGDGQKNLFVASFFFDRAA+AGF +P SPVA VRP DF++AAKQACQTKLE
Sbjct: 184 TFGGIWNGGGGDGQKNLFVASFFFDRAAQAGFANPKSPVAKVRPVDFKNAAKQACQTKLE 363
Query: 399 NAKSTYPRVEEGNLPYLCMDLVYQYTLLVDGFGIYPWQEITLVKKVKYDDALVEAAWPLG 458
+AKSTYP V++GNLPYLCMDLVYQYTLLVDGFG+ P Q+ITLVK+VKY D+LVEAAWPLG
Sbjct: 364 DAKSTYPLVDDGNLPYLCMDLVYQYTLLVDGFGLDPLQDITLVKQVKYHDSLVEAAWPLG 543
Query: 459 SAIEAVSS 466
SAIEAVSS
Sbjct: 544 SAIEAVSS 567
>BI267108 homologue to GP|10717159|gb apyrase 2 {Pisum sativum}, partial
(24%)
Length = 449
Score = 169 bits (428), Expect = 2e-42
Identities = 81/129 (62%), Positives = 105/129 (80%)
Frame = +2
Query: 5 PTRQESLSDKIYRFRGTLLVVSIPILLITFVLYMMPSSSNYDSAGDYALLNRKLSPDKKS 64
P S SDK Y+ RG L++++P+L +T +LY+MPS+S+ +S DYAL +RK+SPD+K
Sbjct: 47 PESSSSFSDKFYQIRGAFLMLALPLLXVTLILYIMPSTSSNESIEDYALTHRKISPDRKI 226
Query: 65 GGSYAVIFDAGSSGSRVHVFHFDQNLDLVHIGKDLELFEQLKPGLSAYAQKPQQAAESLV 124
S+AV+FDAGSSGSRVHVF FD+NL+LV IG DLE+F Q+KPGLSAYA+ PQQAA+SLV
Sbjct: 227 SDSFAVVFDAGSSGSRVHVFRFDRNLELVKIGNDLEVFLQIKPGLSAYARDPQQAAKSLV 406
Query: 125 SLLEKAEGV 133
SLL+KAE V
Sbjct: 407 SLLDKAESV 433
>BF645169 similar to GP|10717159|gb| apyrase 2 {Pisum sativum}, partial (10%)
Length = 542
Score = 64.7 bits (156), Expect = 7e-11
Identities = 31/47 (65%), Positives = 38/47 (79%)
Frame = +1
Query: 154 ALEGDASDKILRAVRDLLKHRSSFKSDADAVTVLDGTQEGAYQWVTI 200
ALEGD SD+IL+AV DLLK RS+ KS+ + V V DGTQEGA+QWV +
Sbjct: 232 ALEGDVSDRILQAV*DLLKQRSTLKSEPNVVDVQDGTQEGAFQWVCL 372
>TC83640 similar to GP|14423550|gb|AAK62457.1 putative nucleoside
triphosphatase {Arabidopsis thaliana}, partial (12%)
Length = 601
Score = 62.0 bits (149), Expect = 5e-10
Identities = 35/95 (36%), Positives = 58/95 (60%)
Frame = +3
Query: 59 SPDKKSGGSYAVIFDAGSSGSRVHVFHFDQNLDLVHIGKDLELFEQLKPGLSAYAQKPQQ 118
SP +KS Y +I D GS+G+RVHVF + + + GK + ++ PGLS++ P
Sbjct: 318 SPPEKSL-RYRIIIDGGSTGTRVHVFKYKMK-NALDFGKKGLVSMRVNPGLSSFGNDPDG 491
Query: 119 AAESLVSLLEKAEGVVPRELRSKTPVRIGATAGLR 153
A SL+ +++ A+ +P+E +T +R+ ATAG+R
Sbjct: 492 AGRSLLEVVDFAKRRIPKENWRETEIRLMATAGMR 596
>AW736308 similar to GP|19920124|gb| Putative nucleoside phosphatase {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (3%)
Length = 552
Score = 61.2 bits (147), Expect = 8e-10
Identities = 46/161 (28%), Positives = 70/161 (42%), Gaps = 15/161 (9%)
Frame = +1
Query: 148 ATAGLRALEGDASDKILRAVRDLLKHRSSFKSDADAVTVLDGTQEGAYQWVTINYLLGNL 207
ATAG+R + D ++L V + K S F D + VL G +E Y WV +NY +G
Sbjct: 7 ATAGMRRIHRDDVFRVLEDVEAVAKDHS-FMFDKRWIRVLSGREEAYYGWVALNYNMGRF 183
Query: 208 GKDY---SKTVGVVDLGGGSVQMAYAISESEAAMAPQVMDGEDPYVKEMFLRGRKYYLYV 264
DY S T+G+VDLGG S+Q+ I G+D ++ +
Sbjct: 184 DDDYHRGSSTLGLVDLGGSSLQIVVEIDRD---------TGDDVNAIRSEFGSIEHRIVA 336
Query: 265 HSYLRYGLLAA------------RAEILKVAGDAENPCILS 293
+S +GL A R E + + +PC++S
Sbjct: 337 YSLPSFGLNEAFDRTVAMLRNNQRVESTRGVSELRHPCLMS 459
>TC81334 weakly similar to GP|19423992|gb|AAL87274.1 putative ABC
transporter protein {Arabidopsis thaliana}, partial
(29%)
Length = 977
Score = 33.9 bits (76), Expect = 0.14
Identities = 24/73 (32%), Positives = 34/73 (45%)
Frame = +3
Query: 25 VSIPILLITFVLYMMPSSSNYDSAGDYALLNRKLSPDKKSGGSYAVIFDAGSSGSRVHVF 84
+SI IL +L++ +S DSA Y ++ R S DKK G ++ A VF
Sbjct: 540 ISIEILTRPRLLFLDEPTSGLDSAASYYVMKRIASLDKKDGIQRTIV--ASIHQPSTEVF 713
Query: 85 HFDQNLDLVHIGK 97
NL L+ GK
Sbjct: 714 QLFHNLCLLSSGK 752
>TC93229 similar to GP|14517424|gb|AAK62602.1 AT5g41370/MYC6_8 {Arabidopsis
thaliana}, partial (19%)
Length = 850
Score = 32.0 bits (71), Expect = 0.52
Identities = 25/79 (31%), Positives = 37/79 (46%), Gaps = 6/79 (7%)
Frame = +1
Query: 65 GGSYAVIFDAGSS--GSRVHVFHFDQNLDL----VHIGKDLELFEQLKPGLSAYAQKPQQ 118
G S+ VI S GSR+H H D L L ++ G D EQL+ A K +
Sbjct: 136 GYSFKVITSLPPSDEGSRLHYHHLDDQLALLGKVLNAGDDAVGLEQLEDDADELALKSAR 315
Query: 119 AAESLVSLLEKAEGVVPRE 137
++ +S + A+G+V E
Sbjct: 316 RSQGSMSAMSGAKGMVYME 372
>TC79258 similar to GP|21592967|gb|AAM64916.1 putative GDSL-motif
lipase/acylhydrolase {Arabidopsis thaliana}, partial
(73%)
Length = 1067
Score = 31.6 bits (70), Expect = 0.68
Identities = 19/43 (44%), Positives = 24/43 (55%)
Frame = +3
Query: 88 QNLDLVHIGKDLELFEQLKPGLSAYAQKPQQAAESLVSLLEKA 130
Q L ++HIGK L+LF Q + LSA Q AE L+ KA
Sbjct: 153 QFLQIIHIGKQLDLFNQYQQKLSA-----QIGAEGAKQLVNKA 266
>TC79897 similar to PIR|E84603|E84603 probable protein transport protein SEC23
[imported] - Arabidopsis thaliana, partial (50%)
Length = 1362
Score = 30.4 bits (67), Expect = 1.5
Identities = 14/31 (45%), Positives = 22/31 (70%)
Frame = -2
Query: 97 KDLELFEQLKPGLSAYAQKPQQAAESLVSLL 127
KDL L E+++PG+ A ++P +A E +SLL
Sbjct: 1139 KDLTLLEEIEPGIHADHKQPGEAQEISLSLL 1047
>TC90584 similar to GP|22136810|gb|AAM91749.1 unknown protein {Arabidopsis
thaliana}, partial (18%)
Length = 686
Score = 30.0 bits (66), Expect = 2.0
Identities = 16/55 (29%), Positives = 27/55 (49%)
Frame = +3
Query: 300 KYGGKSFKASSSGASLNECKSVAHKALKVNESTCTHMKCTFGGIWNGGGGDGQKN 354
K G S +SSS ++ + S + A ++ + T G +GGGGD ++N
Sbjct: 87 KMGPTSSSSSSSSSAASSSSSSSSSAASSSKGSADSSSDTGSGASSGGGGDDEEN 251
>TC93014 similar to PIR|T05880|T05880 hypothetical protein T29A15.240 -
Arabidopsis thaliana, partial (51%)
Length = 758
Score = 30.0 bits (66), Expect = 2.0
Identities = 38/165 (23%), Positives = 68/165 (41%), Gaps = 11/165 (6%)
Frame = +1
Query: 92 LVHIGKDLELFEQLKPGLSAYAQKPQQAAESLVSLLEKAEGVVP-----RELRSKTPVRI 146
L+ I L++ KP S + +Q + S + VV ++ R V +
Sbjct: 88 LISISSRLQIMYLKKPLWSEGIETTKQDSTGSSSSSDPNASVVELVNSLQQQRVYREVTL 267
Query: 147 GATAGLRALEGDASDKILRAVRDLLKHRSSFKSDADAVTVLDGTQEGAYQW---VTINYL 203
T GLR + S LRA+R +L +S + + + TQ + V +
Sbjct: 268 ALTTGLRDARAEFSFLRLRALRSILNFLNSIAHSDSTIYLFNLTQSIPHLQVLPVLFQHS 447
Query: 204 LGNLGKDYS-KTVGVVD--LGGGSVQMAYAISESEAAMAPQVMDG 245
L G DY+ VG + G +++ ++ E A+A +V++G
Sbjct: 448 LKETGNDYNYSRVGDMSHIFGVEPMKLTSPSTDDEIALALRVLEG 582
>TC83122 similar to PIR|C84658|C84658 3-beta-hydroxysteroid dehydrogenase
[imported] - Arabidopsis thaliana, partial (76%)
Length = 1102
Score = 29.6 bits (65), Expect = 2.6
Identities = 20/50 (40%), Positives = 22/50 (44%), Gaps = 9/50 (18%)
Frame = +3
Query: 314 SLNECKSVAHKALKVNESTCTHMKCTFGG--------IWN-GGGGDGQKN 354
SLN +S KA + C H CTFGG IWN G D KN
Sbjct: 618 SLNFGRSWISKAKHKDPCLCYHANCTFGGVDI*AAWPIWNEGASADPFKN 767
>TC79322 similar to GP|13605815|gb|AAK32893.1 At2g32700/F24L7.16
{Arabidopsis thaliana}, partial (59%)
Length = 1824
Score = 28.9 bits (63), Expect = 4.4
Identities = 18/54 (33%), Positives = 23/54 (42%), Gaps = 5/54 (9%)
Frame = +1
Query: 299 YKYGGKSFKASSSGASLNECKSVAHKALKVN-----ESTCTHMKCTFGGIWNGG 347
++ F SSS AS +ECKSV+ A TCT F +W G
Sbjct: 37 HRTNSPGFWCSSSKASPSECKSVSAFATTTTAAAPCSGTCTRKYWQFTSVWRYG 198
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.135 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,514,613
Number of Sequences: 36976
Number of extensions: 161056
Number of successful extensions: 902
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 863
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 881
length of query: 467
length of database: 9,014,727
effective HSP length: 100
effective length of query: 367
effective length of database: 5,317,127
effective search space: 1951385609
effective search space used: 1951385609
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC123593.7


