

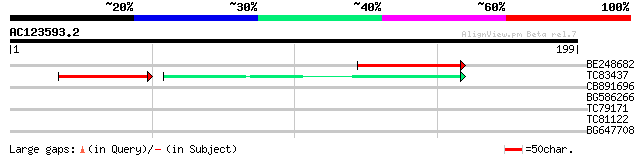
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123593.2 + phase: 0 /pseudo
(199 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BE248682 similar to GP|18568269|gb putative gag-pol polyprotein ... 60 4e-10
TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imp... 32 9e-05
CB891696 35 0.025
BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-li... 33 0.095
TC79171 similar to GP|19310435|gb|AAL84954.1 F17H15.1/F17H15.1 {... 28 1.8
TC81122 similar to GP|20258999|gb|AAM14215.1 unknown protein {Ar... 27 5.2
BG647708 weakly similar to GP|13786450|gb| putative reverse tran... 26 8.9
>BE248682 similar to GP|18568269|gb putative gag-pol polyprotein {Zea mays},
partial (1%)
Length = 441
Score = 60.5 bits (145), Expect = 4e-10
Identities = 29/38 (76%), Positives = 33/38 (86%)
Frame = +3
Query: 123 YADDTLCIGETTVENLYTLKAMLREFEMASSLKVNFLK 160
YADDTLCIG TV+NL+TLKA+L+ FEMAS LKVNF K
Sbjct: 174 YADDTLCIGMPTVDNLWTLKALLQGFEMASGLKVNFHK 287
>TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 951
Score = 31.6 bits (70), Expect(2) = 9e-05
Identities = 32/106 (30%), Positives = 41/106 (38%)
Frame = +2
Query: 55 WIKDRVFGGNVSILVNGSPTEEISKQGDHLTPFLCSRRF**VDVQCGESK*V*GF*GC*R 114
WIK+ V S+LVNGSPT + K T F V+
Sbjct: 407 WIKECVSTATTSVLVNGSPTNVLMKSLVQ-TQLFTRYSFGVVNPVVVSH----------- 550
Query: 115 RDGDFTSSYADDTLCIGETTVENLYTLKAMLREFEMASSLKVNFLK 160
+A+DTL + N+ L+A L F S LKVNF K
Sbjct: 551 ------LQFANDTLLLETKNWANIRALRAALVIF*AMSGLKVNFHK 670
Score = 30.4 bits (67), Expect(2) = 9e-05
Identities = 20/33 (60%), Positives = 21/33 (63%)
Frame = +3
Query: 18 *GSGFG*EEET*TFDF*SGF*KGL*FGRLGLCG 50
*G G G*E E +F * GF*KGL RLGL G
Sbjct: 264 *GCG*G*ETEERSFVV*GGF*KGLSLCRLGLFG 362
>CB891696
Length = 638
Score = 34.7 bits (78), Expect = 0.025
Identities = 16/26 (61%), Positives = 21/26 (80%)
Frame = +1
Query: 135 VENLYTLKAMLREFEMASSLKVNFLK 160
VEN+ T+K ++ FE+ASSL VNFLK
Sbjct: 4 VENILTMKTIVSYFELASSLWVNFLK 81
>BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 789
Score = 32.7 bits (73), Expect = 0.095
Identities = 18/49 (36%), Positives = 26/49 (52%), Gaps = 5/49 (10%)
Frame = -3
Query: 45 RLGLCGKRVGWIKDRVFGGNVSILVNGSPTEEI-----SKQGDHLTPFL 88
RLG G + WI + V + S L+NG P + +QGD L+P+L
Sbjct: 496 RLGFHGIWISWIMECVSTVSYSFLINGGPQGRVLPSRGLRQGDPLSPYL 350
>TC79171 similar to GP|19310435|gb|AAL84954.1 F17H15.1/F17H15.1 {Arabidopsis
thaliana}, partial (27%)
Length = 1421
Score = 28.5 bits (62), Expect = 1.8
Identities = 11/29 (37%), Positives = 19/29 (64%)
Frame = -2
Query: 44 GRLGLCGKRVGWIKDRVFGGNVSILVNGS 72
GRLG G R+ W++ + G++++L N S
Sbjct: 859 GRLGRRGYRITWLRSSLLVGSIALLGNSS 773
>TC81122 similar to GP|20258999|gb|AAM14215.1 unknown protein {Arabidopsis
thaliana}, partial (61%)
Length = 675
Score = 26.9 bits (58), Expect = 5.2
Identities = 10/23 (43%), Positives = 17/23 (73%)
Frame = -3
Query: 177 VISFIIVKGQSHLLIWIFQLVQI 199
++SF+++ S+L IWIF L+ I
Sbjct: 466 IVSFVLLNKLSNLFIWIFLLIII 398
>BG647708 weakly similar to GP|13786450|gb| putative reverse transcriptase
{Oryza sativa}, partial (9%)
Length = 708
Score = 26.2 bits (56), Expect = 8.9
Identities = 12/38 (31%), Positives = 19/38 (49%)
Frame = +1
Query: 123 YADDTLCIGETTVENLYTLKAMLREFEMASSLKVNFLK 160
+ADD+L + T+ +L ++ AS VNF K
Sbjct: 160 FADDSLLFARANLTEAATIMQVLHSYQSASGQLVNFEK 273
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.363 0.169 0.619
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,580,070
Number of Sequences: 36976
Number of extensions: 67941
Number of successful extensions: 638
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 634
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 638
length of query: 199
length of database: 9,014,727
effective HSP length: 91
effective length of query: 108
effective length of database: 5,649,911
effective search space: 610190388
effective search space used: 610190388
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (22.0 bits)
S2: 56 (26.2 bits)
Medicago: description of AC123593.2


