

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123574.2 + phase: 0 /pseudo
(984 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
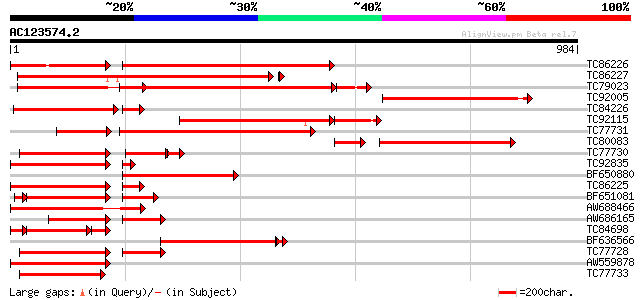
Score E
Sequences producing significant alignments: (bits) Value
TC86226 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 433 0.0
TC86227 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 536 e-152
TC79023 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 437 e-134
TC92005 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 437 e-123
TC84226 similar to GP|22037313|gb|AAM89998.1 disease resistance-... 374 e-122
TC92115 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 338 e-114
TC77731 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 379 e-105
TC80083 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 249 2e-85
TC77730 weakly similar to GP|7341113|gb|AAF61210.1| unknown {Gly... 232 4e-81
TC92835 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 280 3e-80
BF650880 weakly similar to GP|9858476|gb| resistance protein LM1... 263 2e-70
TC86225 weakly similar to GP|9858476|gb|AAG01051.1| resistance p... 261 9e-70
BF651081 weakly similar to GP|18033111|g functional candidate re... 229 1e-66
AW688466 similar to GP|9965109|gb| resistance protein MG13 {Glyc... 249 3e-66
AW686165 weakly similar to GP|18033111|gb functional candidate r... 174 9e-65
TC84698 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 187 2e-64
BF636566 weakly similar to GP|9965103|gb| resistance protein LM6... 234 5e-64
TC77728 similar to GP|12056928|gb|AAG48132.1 putative resistance... 238 8e-63
AW559878 weakly similar to GP|18033111|g functional candidate re... 237 1e-62
TC77733 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 236 3e-62
>TC86226 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (24%)
Length = 1769
Score = 433 bits (1114), Expect(2) = 0.0
Identities = 248/367 (67%), Positives = 280/367 (75%)
Frame = +2
Query: 197 I*I*FYWRHSQIHLQQDQSCALTCCQLPCWI*VSNTTSEIAS**GN**RGTHGRTLWDRR 256
I*I YW+ SQIHLQQDQS LTCC LPCWI VS+TTSEIAS**G ** G +G LWD R
Sbjct: 542 I*IRAYWKDSQIHLQQDQSTVLTCCHLPCWIAVSSTTSEIAS**GT**WGPYGWNLWDWR 721
Query: 257 LG*INTCKSNL*FYW*SI*RIMFSS*CEREFS*K*LETSPREAPFENNWIGN*VRPC**G 316
L *INTCKSNL*F *SI*R MFS +REFS K*LET P +APF++ N*V C *G
Sbjct: 722 LR*INTCKSNL*FCC*SI*RFMFS*TSQREFSFK*LETFPGDAPFKDTSTEN*VSRCQ*G 901
Query: 317 NSNH*GKAM*KEDSFDSR*C*QYESTACFGWRT*LVRPWKQSHHYHSRQTLTK*SWDKKY 376
N NH GK +*KEDSFDSR* *QYE+T CFGWR+*LVR WK SHH+H R TLT S D+K
Sbjct: 902 NLNHKGKVV*KEDSFDSR*R*QYEATECFGWRS*LVRSWKPSHHHHPR*TLTSLS*DRKN 1081
Query: 377 TCSRRVEWD*SS*IVKVDGF*K*QSSFRLRRHFKPCSCIFFWPSISYRSSGFKLVWKEHR 436
CS+RVE D SS*IV+VDGF*K*QSSF+L + F+ CSC+ FWPS SYR+SGF LVWKEHR
Sbjct: 1082ICSKRVECDRSS*IVEVDGF*K*QSSFKL*KDFESCSCLCFWPSGSYRNSGF*LVWKEHR 1261
Query: 437 KMEEYIRWV*QDSQ*RDPKDT*SKL*CFGRRGAECLSGHCLLLQRMRMGGCQRYTPCSLW 496
+M+EYIR V*+DSQ*RDPK+T*S+L* FG RGA+CLS HCLLLQR+ MG QR T CSLW
Sbjct: 1262RMQEYIRLV*KDSQ*RDPKNT*SEL*FFGGRGAKCLSRHCLLLQRL*MGKGQRNTSCSLW 1441
Query: 497 PLHNTSSRSVG*KISHRPLGI*WLCGIT*LDRGHG*RSRPTRIT*RARRKK*AMVSR*YS 556
LH +S S *K+SH P I C IT*LDR HG*R+RPTRIT A +KK MV + Y
Sbjct: 1442SLHKSSC*SAS*KMSH*PF*IR*SCVIT*LDRKHG*RTRPTRITF*AWKKKQVMV*KGYI 1621
Query: 557 SCFK*KH 563
+ KH
Sbjct: 1622*SSRRKH 1642
Score = 226 bits (575), Expect(2) = 0.0
Identities = 119/178 (66%), Positives = 137/178 (76%), Gaps = 3/178 (1%)
Frame = +3
Query: 1 MQSSSSSSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDD--SELQRGDE 58
MQ SSS SY F Y VFL +G+DTRYGFTGNL KAL DKGI TF DD S+LQR D+
Sbjct: 45 MQLPFYSSSFSYAFSYDVFLICKGTDTRYGFTGNLLKALIDKGIRTFHDDDDSDLQRRDK 224
Query: 59 ITPSLDNAIEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVR 118
+TP + IEESRI IP+FSANYASSS CLD LVHIIH YK G LVLPVFFGV+P+ VR
Sbjct: 225 VTPII---IEESRILIPIFSANYASSSSCLDTLVHIIHCYKTKGCLVLPVFFGVEPTDVR 395
Query: 119 HHRGSYGEALAKHEERFQHNTDHMERLQKWKIALTQAANLSGDH-RSPGYEYKLTGKI 175
HH G YG+ALA+HE RFQ++T +MERLQ+WK+AL+ AANL H S GYEY+L GKI
Sbjct: 396 HHTGRYGKALAEHENRFQNDTKNMERLQQWKVALSLAANLPSYHDDSHGYEYELIGKI 569
>TC86227 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (22%)
Length = 1736
Score = 536 bits (1382), Expect(2) = e-152
Identities = 301/485 (62%), Positives = 344/485 (70%), Gaps = 40/485 (8%)
Frame = +3
Query: 14 FKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIF 73
FKY VFLSFR DT YGFTGNLYKAL DKGI TFIDD++L+RGDE TPSL AIEESRI
Sbjct: 12 FKYHVFLSFRDIDTLYGFTGNLYKALIDKGIKTFIDDNDLERGDESTPSLVKAIEESRIL 191
Query: 74 IPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEALAKHEE 133
IP+FSANYASSSFCLDELVHIIH YK G VLPVF+G DP+HVRH GSYGE L KHE+
Sbjct: 192 IPIFSANYASSSFCLDELVHIIHCYKTRGCSVLPVFYGADPTHVRHQTGSYGEHLTKHED 371
Query: 134 RFQHNTDHMERLQKWKIALTQAANLSGDHRSPGYE---------YKLTGKIAFNQTPDLS 184
+FQ+N ++MERL+KWK+ALTQAAN SG H S GY Y I+F L
Sbjct: 372 KFQNNKENMERLKKWKMALTQAANFSGHHFSQGYPTLLNIIFPFYIFFSLISFPH*SHLK 551
Query: 185 S-------------------------------DCSQSLVEHEQI*I*FYWRHSQIHLQQD 213
+ D + + HE I*+ Y +S+ H + D
Sbjct: 552 A*KGSMFFIYVFKQVYVSKQLELYNILSIHGCDFTNLSIGHE*I*VRAY*EYSRTHFR*D 731
Query: 214 QSCALTCCQLPCWI*VSNTTSEIAS**GN**RGTHGRTLWDRRLG*INTCKSNL*FYW*S 273
QSC T CQ+PCWI V +TTS+IAS *G **RG HGRTLW LG INTCKSNL*F+ *S
Sbjct: 732 QSCFSTRCQIPCWITVPSTTSKIASG*GI**RGQHGRTLWY*SLGKINTCKSNL*FHC*S 911
Query: 274 I*RIMFSS*CEREFS*K*LETSPREAPFENNWIGN*VRPC**GNSNH*GKAM*KEDSFDS 333
I*R M SS*C REFS LETSP APF+N+ +*V+ C *GNSNH*GKAM*+EDSFDS
Sbjct: 912 I*RSMLSS*C*REFSS*KLETSPEGAPFKNSSTEH*VKRCF*GNSNH*GKAM*EEDSFDS 1091
Query: 334 R*C*QYESTACFGWRT*LVRPWKQSHHYHSRQTLTK*SWDKKYTCSRRVEWD*SS*IVKV 393
R*C* S FGWRT*LVR WKQSHH+H R TLT SWD+K CS+R W+ SS*IV+V
Sbjct: 1092R*C*PTGSARGFGWRT*LVRSWKQSHHHHPR*TLTNLSWDRKNICSKRAIWEGSS*IVEV 1271
Query: 394 DGF*K*QSSFRLRRHFKPCSCIFFWPSISYRSSGFKLVWKEHRKMEEYIRWV*QDSQ*RD 453
DGF*+*QS+ +L R FKPCS I+F S+S+R+SGF LVWKEHR MEE+IRWV*QDSQ*RD
Sbjct: 1272DGF*E*QSTAKL*RCFKPCSFIWFGHSLSFRNSGF*LVWKEHRSMEEHIRWV*QDSQ*RD 1451
Query: 454 PKDT* 458
KDT*
Sbjct: 1452TKDT* 1466
Score = 23.1 bits (48), Expect(2) = e-152
Identities = 8/11 (72%), Positives = 10/11 (90%)
Frame = +1
Query: 467 RGAECLSGHCL 477
RGA+C+S HCL
Sbjct: 1465 RGAKCIS*HCL 1497
>TC79023 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (37%)
Length = 2072
Score = 437 bits (1125), Expect(2) = e-134
Identities = 249/376 (66%), Positives = 283/376 (75%)
Frame = +2
Query: 191 LVEHEQI*I*FYWRHSQIHLQQDQSCALTCCQLPCWI*VSNTTSEIAS**GN**RGTHGR 250
L+ HE I*I YW+ S+ H QQD S AL CCQ+ CWI V +TT E AS** ** G HGR
Sbjct: 671 LIGHE*I*IRVYWKDSRRHFQQD*S*ALRCCQISCWITVPSTTCERAS**KI***GPHGR 850
Query: 251 TLWDRRLG*INTCKSNL*FYW*SI*RIMFSS*CEREFS*K*LETSPREAPFENNWIGN*V 310
TLW R *INTCKSNL*F+ *SI* MFS CE EF +*LETSPRE PF+N+ IG+*V
Sbjct: 851 TLWYWRHR*INTCKSNL*FHC*SI*SFMFSRKCESEFDFR*LETSPRETPFKNS*IGH*V 1030
Query: 311 RPC**GNSNH*GKAM*KEDSFDSR*C*QYESTACFGWRT*LVRPWKQSHHYHSRQTLTK* 370
R C* GNSNH* KAM*KEDSFDS *C*Q S GWRT*LVR W+ SHHY+S+QTLTK
Sbjct: 1031 RRC*SGNSNH*AKAM*KEDSFDS**C*QTGSARGLGWRT*LVRSWE*SHHYYSKQTLTKD 1210
Query: 371 SWDKKYTCSRRVEWD*SS*IVKVDGF*K*QSSFRLRRHFKPCSCIFFWPSISYRSSGFKL 430
SWD+KYTCSRRVE D SS IV+VDGF + + SF+ RHFKPC+ + FWPSISY ++ +
Sbjct: 1211 SWDRKYTCSRRVECDRSSRIVEVDGFQR-KCSFKS*RHFKPCTYLCFWPSISYCNNRVQF 1387
Query: 431 VWKEHRKMEEYIRWV*QDSQ*RDPKDT*SKL*CFGRRGAECLSGHCLLLQRMRMGGCQRY 490
WKE + EYIRWV*++SQ*RDPK+T*S+L* FG R A+CLS HCLLLQR+ M QR
Sbjct: 1388 GWKERTRFNEYIRWV*RNSQ*RDPKNT*SEL*FFGERRAKCLSRHCLLLQRV*MARGQRN 1567
Query: 491 TPCSLWPLHNTSSRSVG*KISHRPLGI*WLCGIT*LDRGHG*RSRPTRIT*RARRKK*AM 550
T CSLW LH+TS S G*KISH P I* LC IT*LDRGHG*RSRPTRIT A R +* +
Sbjct: 1568 TSCSLWSLHSTSCCSFG*KISHGPFEI**LCDIT*LDRGHG*RSRPTRITR*AWRTQ*VV 1747
Query: 551 VSR*YSSCFK*KHYQK 566
V *YSSCFK KH+ K
Sbjct: 1748 V*A*YSSCFKKKHWNK 1795
Score = 261 bits (667), Expect = 9e-70
Identities = 136/225 (60%), Positives = 158/225 (69%)
Frame = +1
Query: 14 FKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIF 73
F YQVFLSFRG+DTR+GFTGNLYKALTDKGI+TFIDD++LQRGDEITPSL NAIE+SRIF
Sbjct: 88 FTYQVFLSFRGADTRHGFTGNLYKALTDKGIYTFIDDNDLQRGDEITPSLKNAIEKSRIF 267
Query: 74 IPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEALAKHEE 133
IPVFS NYASSSFCLDELVHI H Y G LVLPVF GVDP+ VRHH G YGEALA H++
Sbjct: 268 IPVFSENYASSSFCLDELVHITHCYDTKGCLVLPVFIGVDPTDVRHHTGRYGEALAVHKK 447
Query: 134 RFQHNTDHMERLQKWKIALTQAANLSGDHRSPGYEYKLTGKIAFNQTPDLSSDCSQSLVE 193
+FQ++ D+ ERLQ+WK AL+QAANLSG H GY ++
Sbjct: 448 KFQNDKDNTERLQQWKEALSQAANLSGQHYKHGYRIRV---------------------- 561
Query: 194 HEQI*I*FYWRHSQIHLQQDQSCALTCCQLPCWI*VSNTTSEIAS 238
YW+ S+ H QQD S A C Q+PC I +TT + AS
Sbjct: 562 --------YWKDSRRHFQQD*SQAPRCSQIPC*ITAPSTTYKNAS 672
Score = 59.7 bits (143), Expect(2) = e-134
Identities = 35/65 (53%), Positives = 43/65 (65%), Gaps = 3/65 (4%)
Frame = +3
Query: 567 GMAFKKMTKLKTLIIENGHFSNGLKYLPNSLRVLKWKGCLLESLSSSILSKASE---VTS 623
G AF+KMT LKT I ENGH S L+YLP+SLRV+ KGC+ +S SSS +K E V
Sbjct: 1854 GNAFEKMTNLKTFITENGHHSKSLEYLPSSLRVM--KGCIPKSPSSSSSNKKFEDMKVLI 2027
Query: 624 FYNCK 628
NC+
Sbjct: 2028 LNNCE 2042
Score = 30.4 bits (67), Expect = 3.5
Identities = 18/24 (75%), Positives = 20/24 (83%)
Frame = +1
Query: 642 RSSRI*KS*HWTIVNI*HIYLMSQ 665
RS +I*KS*+ TIVNI* IY MSQ
Sbjct: 1999 RSLKI*KS*Y*TIVNI*PIYRMSQ 2070
>TC92005 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (7%)
Length = 858
Score = 437 bits (1124), Expect = e-123
Identities = 238/260 (91%), Positives = 245/260 (93%)
Frame = +2
Query: 648 KS*HWTIVNI*HIYLMSQVFQI*RSSHLNFVEI*LQLTIQLATKINLNS*VQLVAASSSV 707
KS*HWTIVNI*HIYLMSQVFQI*RSSHLNFVEI*LQLTIQLATKINLNS*VQLVAASSSV
Sbjct: 2 KS*HWTIVNI*HIYLMSQVFQI*RSSHLNFVEI*LQLTIQLATKINLNS*VQLVAASSSV 181
Query: 708 FHP*VWHLLRNWNSLFV*VSIVFQNYYAR*QI*NVFCLLILP*ENCHLHFKISVNLMTYQ 767
FHP*VWHLLRNWNSLFV*VSIVFQNYYAR*QI*NVFCLLILP*ENCHLHFKISVNLMTYQ
Sbjct: 182 FHP*VWHLLRNWNSLFV*VSIVFQNYYAR*QI*NVFCLLILP*ENCHLHFKISVNLMTYQ 361
Query: 768 *RDVEC*GSQNIMTKLIL*CFQM*HSLVSKIATSQMNVYQYFSSGLLM*KG*TYRITSIS 827
*RDVEC*GSQNIMTKLIL*CFQM*HSLVSKIATSQMNVYQYFSSGLLM*KG*TYRITSIS
Sbjct: 362 *RDVEC*GSQNIMTKLIL*CFQM*HSLVSKIATSQMNVYQYFSSGLLM*KG*TYRITSIS 541
Query: 828 IFFPSVLMNVTL*RSLNSIVASLLRKLEGFHQILKSFPLINVNR*VPQVEEC*RVSLKLE 887
IFFPSVLMNVTL*RSLNSIVASLLRKLEGFHQILKSFPLINVNR*VPQVEEC*RV
Sbjct: 542 IFFPSVLMNVTL*RSLNSIVASLLRKLEGFHQILKSFPLINVNR*VPQVEEC*RV----- 706
Query: 888 EHNKRLRRIYGIKYESEMDK 907
+ R+ + + S+M++
Sbjct: 707 ---RNYMRLEALNFISQMEQ 757
>TC84226 similar to GP|22037313|gb|AAM89998.1 disease resistance-like
protein GS0-1 {Glycine max}, partial (72%)
Length = 665
Score = 374 bits (960), Expect(2) = e-122
Identities = 182/183 (99%), Positives = 182/183 (99%)
Frame = +3
Query: 7 SSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNA 66
SSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNA
Sbjct: 3 SSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNA 182
Query: 67 IEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGE 126
IEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGE
Sbjct: 183 IEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGE 362
Query: 127 ALAKHEERFQHNTDHMERLQKWKIALTQAANLSGDHRSPGYEYKLTGKIAFNQTPDLSSD 186
ALAKHEERFQHNTDHMERLQKWKIALTQAANLSGDHRSPGYEYKLTGKIAFNQTP LSSD
Sbjct: 363 ALAKHEERFQHNTDHMERLQKWKIALTQAANLSGDHRSPGYEYKLTGKIAFNQTPHLSSD 542
Query: 187 CSQ 189
CSQ
Sbjct: 543 CSQ 551
Score = 83.2 bits (204), Expect(2) = e-122
Identities = 37/38 (97%), Positives = 38/38 (99%)
Frame = +2
Query: 196 QI*I*FYWRHSQIHLQQDQSCALTCCQLPCWI*VSNTT 233
+I*I*FYWRHSQIHLQQDQSCALTCCQLPCWI*VSNTT
Sbjct: 551 EI*I*FYWRHSQIHLQQDQSCALTCCQLPCWI*VSNTT 664
>TC92115 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (24%)
Length = 1348
Score = 338 bits (867), Expect(2) = e-114
Identities = 186/274 (67%), Positives = 210/274 (75%), Gaps = 4/274 (1%)
Frame = +1
Query: 295 SPREAPFENNWIGN*VRPC**GNSNH*GKAM*KEDSFDSR*C*QYESTACFGWRT*LVRP 354
SP APF++ N*VR C**GNS + GKA KEDSFDSR*C*+Y +T CFG RT*LVRP
Sbjct: 1 SPGGAPFKSTSTEN*VRRC**GNSLYKGKAAYKEDSFDSR*C*RYGATTCFGRRT*LVRP 180
Query: 355 WKQSHHYHSRQTLTK*SWDKKYTCSRRVEWD*SS*IVKVDGF*K*QSSFRLRRHFKPCSC 414
WKQ HHYHSRQTLTK SWD+KY S R WD SS +V++DGF*+*QS+F L R FK CS
Sbjct: 181 WKQGHHYHSRQTLTKKSWDRKYA*SGRAVWDRSSRVVEMDGF*E*QSTFNL*RRFKSCSF 360
Query: 415 IFFWPSISYRSSGFKLVWKEHRKMEEYIRWV*QDSQ*RDPKDT*SKL*CFGRRGAECLSG 474
I F PS+S+R+SGF+LVWK+ R+ME YIRW+*+DSQ*+DP DT*SKL*CFG R A+C S
Sbjct: 361 ICFGPSLSFRNSGFQLVWKDDRRMEGYIRWI*KDSQ*KDPPDT*SKL*CFGGRAAKCFSR 540
Query: 475 HCLLLQRMRMGGCQRYTPCSLWPLHNTSSRSVG*KISH----RPLGI*WLCGIT*LDRGH 530
HCLLLQRM MGG YT CSLWP HNTSS SVG*KIS PL I T*LD+G+
Sbjct: 541 HCLLLQRMWMGGV*IYTSCSLWPSHNTSSSSVG*KISR*DYSSPLWIYL*TDTT*LDKGN 720
Query: 531 G*RSRPTRIT*RARRKK*AMVSR*YSSCFK*KHY 564
G*RSRPTRI RA RKK* MV R*YS CFK KH+
Sbjct: 721 G*RSRPTRIAQRAWRKK*IMV*R*YS*CFKRKHW 822
Score = 92.8 bits (229), Expect(2) = e-114
Identities = 50/81 (61%), Positives = 58/81 (70%)
Frame = +2
Query: 565 QKGMAFKKMTKLKTLIIENGHFSNGLKYLPNSLRVLKWKGCLLESLSSSILSKASEVTSF 624
+KG AFKKMT+LKTLIIEN HFS GLKYLP+SLRVLK +GCL ESL S LSKA E+TSF
Sbjct: 881 KKGKAFKKMTRLKTLIIENVHFSKGLKYLPSSLRVLKLRGCLSESLISCSLSKAIEITSF 1060
Query: 625 YNCKITKRVFVSSLCLRRSSR 645
C + + C R+ R
Sbjct: 1061SYC--IYLLIYNDFCFRQQRR 1117
Score = 41.2 bits (95), Expect = 0.002
Identities = 31/66 (46%), Positives = 39/66 (58%), Gaps = 6/66 (9%)
Frame = +1
Query: 633 VFVSSLCLRRSSRI*KS*HWTIVNI*HIYLMSQVFQI*RSSHLNFVEI*L------QLTI 686
+FVSSL L RSSR *KS*HWT VNI ++ + + RSS V +*+ Q TI
Sbjct: 1156 LFVSSLXLCRSSRT*KS*HWTGVNI-----LTHIPDVLRSSKFKKVLL*ILXEFNYQFTI 1320
Query: 687 QLATKI 692
QL T +
Sbjct: 1321 QLDTSL 1338
>TC77731 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (23%)
Length = 1651
Score = 379 bits (974), Expect = e-105
Identities = 222/341 (65%), Positives = 249/341 (72%)
Frame = +3
Query: 191 LVEHEQI*I*FYWRHSQIHLQQDQSCALTCCQLPCWI*VSNTTSEIAS**GN**RGTHGR 250
L+ HE I*I Y S+ HLQQ QSC Q+PCWI V N TSEIAS* G * G HGR
Sbjct: 480 LIGHE*I*IQVY*EDSRRHLQQYQSCFFKRSQIPCWITVPNRTSEIAS*HGL*G*GPHGR 659
Query: 251 TLWDRRLG*INTCKSNL*FYW*SI*RIMFSS*CEREFS*K*LETSPREAPFENNWIGN*V 310
T W R G*INTCKS+L*F *SI*R MFSS*C+REF+ ETSP+EAPF+N+ I *V
Sbjct: 660 TFWYWRHG*INTCKSSL*FCC*SI*RCMFSS*CQREFNS*EPETSPKEAPFKNS*I*R*V 839
Query: 311 RPC**GNSNH*GKAM*KEDSFDSR*C*QYESTACFGWRT*LVRPWKQSHHYHSRQTLTK* 370
R C**GN+NH GK +*KEDSFDS *C*Q + FGWRT*LVR WKQ HHYHS TLT
Sbjct: 840 RRC**GNTNHKGKTI*KEDSFDS**C*QTGAARGFGWRT*LVRSWKQGHHYHS**TLTSL 1019
Query: 371 SWDKKYTCSRRVEWD*SS*IVKVDGF*K*QSSFRLRRHFKPCSCIFFWPSISYRSSGFKL 430
SWD KYTCSRR+E D SS*IV+ +GF*K*QSSF L R FKPCS + W ++S ++
Sbjct: 1020SWDYKYTCSRRIE*DRSS*IVEANGF*K*QSSFNL*RDFKPCSYLCIWAALSNSNNRG*F 1199
Query: 431 VWKEHRKMEEYIRWV*QDSQ*RDPKDT*SKL*CFGRRGAECLSGHCLLLQRMRMGGCQRY 490
VWKE R++E YIR V*+ SQ*R PKDT S L*CFG +G ECLS HCLLLQR+ M +
Sbjct: 1200VWKEGRRLEAYIR*V*KYSQ*RYPKDTSS*L*CFGTKGEECLSRHCLLLQRV*MDKG*KN 1379
Query: 491 TPCSLWPLHNTSSRSVG*KISHRPLGI*WLCGIT*LDRGHG 531
T CSLWPLH TS SVG*KISHRPLGI* +T*LDRGHG
Sbjct: 1380TSCSLWPLHRTSCWSVG*KISHRPLGI*HSDDLT*LDRGHG 1502
Score = 135 bits (340), Expect = 8e-32
Identities = 67/98 (68%), Positives = 76/98 (77%), Gaps = 1/98 (1%)
Frame = +2
Query: 81 YASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEALAKHEERFQHNTD 140
YASSSFCLDELVHIIH YK LVLPVF+ V+P+H+RH GSYGE L KHEERFQ+N
Sbjct: 2 YASSSFCLDELVHIIHCYKTKSCLVLPVFYDVEPTHIRHQSGSYGEYLTKHEERFQNNEK 181
Query: 141 HMERLQKWKIALTQAANLSGDHRSP-GYEYKLTGKIAF 177
+MERL++WKIALTQAANLSG H SP GY L +F
Sbjct: 182 NMERLRQWKIALTQAANLSGYHYSPHGYPTLLKTSFSF 295
Score = 38.1 bits (87), Expect = 0.017
Identities = 23/37 (62%), Positives = 27/37 (72%), Gaps = 3/37 (8%)
Frame = +1
Query: 633 VFVSSLCL---RRSSRI*KS*HWTIVNI*HIYLMSQV 666
+ +SSLCL RSS I*KS*+W IV I* IY MS+V
Sbjct: 1540 IMISSLCLVF*ARSSTI*KS*NWIIVQI*PIYPMSRV 1650
>TC80083 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (4%)
Length = 1269
Score = 249 bits (636), Expect(2) = 2e-85
Identities = 155/237 (65%), Positives = 177/237 (74%)
Frame = +1
Query: 642 RSSRI*KS*HWTIVNI*HIYLMSQVFQI*RSSHLNFVEI*LQLTIQLATKINLNS*VQLV 701
RSSRI*KS*+WT VNI*HIY MSQ FQI*+S L V I*LQ TI+L T INL *V +V
Sbjct: 235 RSSRI*KS*YWTDVNI*HIYPMSQAFQI*KSYRLKDVII*LQFTIRLDTLINLKG*VHMV 414
Query: 702 AASSSVFHP*VWHLLRNWNSLFV*VSIVFQNYYAR*QI*NVFCLLILP*ENCHLHFKISV 761
A S++FH *VWHLLRN S+ VS VFQNYYAR*QI* +F +ILP*ENCH HFKI V
Sbjct: 415 AIRSNIFHL*VWHLLRN*ISVLAGVSRVFQNYYAR*QI*TIFGCVILP*ENCHFHFKILV 594
Query: 762 NLMTYQ*RDVEC*GSQNIMTKLIL*CFQM*HSLVSKIATSQMNVYQYFSSGLLM*KG*TY 821
N ++YQ* +EC*GSQN M + I *CFQM*HSL+S IATSQMNV + FSSG+L+* *TY
Sbjct: 595 NFISYQ*H-MEC*GSQNRMIRCIQ*CFQM*HSLLSTIATSQMNVSEEFSSGVLI*HL*TY 771
Query: 822 RITSISIFFPSVLMNVTL*RSLNSIVASLLRKLEGFHQILKSFPLINVNR*VPQVEE 878
I +IS FF S L+NVT+ L AS LRKLEGFHQI K +VN *VPQV+E
Sbjct: 772 YI-AISKFFLSALVNVTILLKLMWGTASPLRKLEGFHQI*KRLMHKDVNH*VPQVKE 939
Score = 86.3 bits (212), Expect(2) = 2e-85
Identities = 42/53 (79%), Positives = 46/53 (86%)
Frame = +3
Query: 565 QKGMAFKKMTKLKTLIIENGHFSNGLKYLPNSLRVLKWKGCLLESLSSSILSK 617
+KG AFKKMTKLKTLIIENGH S GLKYLP+SL VLKWKGCL + LSS IL+K
Sbjct: 78 EKGKAFKKMTKLKTLIIENGHCSKGLKYLPSSLSVLKWKGCLSKCLSSRILNK 236
>TC77730 weakly similar to GP|7341113|gb|AAF61210.1| unknown {Glycine max},
partial (80%)
Length = 831
Score = 232 bits (591), Expect(3) = 4e-81
Identities = 118/160 (73%), Positives = 132/160 (81%), Gaps = 1/160 (0%)
Frame = +1
Query: 17 QVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIFIPV 76
+VFL+FRGSDTR FTGNLYKAL DKGI TFID+++LQRGDEITPSL AIEESRI IP+
Sbjct: 37 RVFLNFRGSDTRNNFTGNLYKALVDKGIRTFIDENDLQRGDEITPSLVKAIEESRICIPI 216
Query: 77 FSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEALAKHEERFQ 136
FSANYASSSFCLDELVHIIH YK LVLPVF+GVDP+ VR H SYGE L KHEE FQ
Sbjct: 217 FSANYASSSFCLDELVHIIHCYKTKSCLVLPVFYGVDPTDVRRHTCSYGEYLTKHEEGFQ 396
Query: 137 HNTDHMERLQKWKIALTQAANLSGDHRSP-GYEYKLTGKI 175
+N +MERL++WK+ALTQAANLSG H SP YE+K KI
Sbjct: 397 NNEKNMERLRQWKMALTQAANLSGYHYSPHEYEHKFIEKI 516
Score = 71.6 bits (174), Expect(3) = 4e-81
Identities = 45/78 (57%), Positives = 48/78 (60%)
Frame = +3
Query: 202 YWRHSQIHLQQDQSCALTCCQLPCWI*VSNTTSEIAS**GN**RGTHGRTLWDRRLG*IN 261
Y S IHLQQ QS Q+PCWI V N TSEIAS* G * G HGRT W R G*IN
Sbjct: 504 Y*EDSPIHLQQYQS*FFKRSQIPCWITVPNRTSEIAS*HGL*G*GPHGRTFWYWRHG*IN 683
Query: 262 TCKSNL*FYW*SI*RIMF 279
TCKS+ *F *S + F
Sbjct: 684 TCKSSF*FSC*SFEGVCF 737
Score = 38.9 bits (89), Expect(3) = 4e-81
Identities = 20/29 (68%), Positives = 24/29 (81%)
Frame = +2
Query: 274 I*RIMFSS*CEREFS*K*LETSPREAPFE 302
I*R MFSS*C+REF * ETSP++APF+
Sbjct: 719 I*RCMFSS*CQREFKS**PETSPKDAPFK 805
>TC92835 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (13%)
Length = 581
Score = 280 bits (717), Expect(2) = 3e-80
Identities = 138/175 (78%), Positives = 151/175 (85%)
Frame = +2
Query: 1 MQSSSSSSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEIT 60
MQ SSSSS S F YQVFLSFRG+DTR+GFTGNLYKALTDKGI TFIDD++LQRGDEIT
Sbjct: 17 MQLPSSSSSFSSAFTYQVFLSFRGTDTRHGFTGNLYKALTDKGIKTFIDDNDLQRGDEIT 196
Query: 61 PSLDNAIEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHH 120
PSL AIEESRIFIPVFS NYA+S FCLDELVHIIH YK GRLVLPVFFGVDP++VRHH
Sbjct: 197 PSLLKAIEESRIFIPVFSINYATSKFCLDELVHIIHCYKTKGRLVLPVFFGVDPTNVRHH 376
Query: 121 RGSYGEALAKHEERFQHNTDHMERLQKWKIALTQAANLSGDHRSPGYEYKLTGKI 175
G YGEALA HE+RFQ++ D+MERL +WK+ALTQAANLSG H SPGYEYK G I
Sbjct: 377 TGPYGEALAGHEKRFQNDKDNMERLHQWKLALTQAANLSGYHSSPGYEYKFIGDI 541
Score = 37.7 bits (86), Expect(2) = 3e-80
Identities = 18/22 (81%), Positives = 19/22 (85%)
Frame = +1
Query: 197 I*I*FYWRHSQIHLQQDQSCAL 218
I*I YWR+SQIHLQQDQS AL
Sbjct: 514 I*IQVYWRYSQIHLQQDQSSAL 579
>BF650880 weakly similar to GP|9858476|gb| resistance protein LM12 {Glycine
max}, partial (41%)
Length = 659
Score = 263 bits (672), Expect = 2e-70
Identities = 147/201 (73%), Positives = 159/201 (78%)
Frame = +2
Query: 197 I*I*FYWRHSQIHLQQDQSCALTCCQLPCWI*VSNTTSEIAS**GN**RGTHGRTLWDRR 256
I*I WR SQIHLQQDQS ALTCC LPCWI +S+T SEIAS**G ** G +G LWD R
Sbjct: 26 I*IQVDWRDSQIHLQQDQSSALTCC*LPCWIALSSTRSEIAS**GT**WGPYGWNLWDWR 205
Query: 257 LG*INTCKSNL*FYW*SI*RIMFSS*CEREFS*K*LETSPREAPFENNWIGN*VRPC**G 316
LG*I+TCKSNL*F *SI*R MFSS*CEREFS K*LETSPREA +NNWI N VR C *G
Sbjct: 206 LG*ISTCKSNL*FCC*SI*RFMFSS*CEREFSSK*LETSPREASSKNNWIENQVRSCL*G 385
Query: 317 NSNH*GKAM*KEDSFDSR*C*QYESTACFGWRT*LVRPWKQSHHYHSRQTLTK*SWDKKY 376
+SN+ GK +*K DSFDSR*C*+Y +T CFGWRT*LV PWKQ HHYHSRQT T S +K
Sbjct: 386 DSNYKGKTV*K*DSFDSR*C*RYGATTCFGWRT*LVWPWKQGHHYHSRQTFTDQS*YRKN 565
Query: 377 TCSRRVEWD*SS*IVKVDGF* 397
CSR V WD SS*IV+ DGF*
Sbjct: 566 ICSRGVVWDKSS*IVEXDGF* 628
>TC86225 weakly similar to GP|9858476|gb|AAG01051.1| resistance protein LM12
{Glycine max}, partial (25%)
Length = 677
Score = 261 bits (667), Expect = 9e-70
Identities = 130/175 (74%), Positives = 144/175 (82%)
Frame = +3
Query: 1 MQSSSSSSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEIT 60
MQS SSSSSIS FKY VFLSFR DT YGFTGNLYKAL DKGI TFIDD++L+RGDE T
Sbjct: 66 MQSLSSSSSISSGFKYHVFLSFRDIDTLYGFTGNLYKALIDKGIKTFIDDNDLERGDEST 245
Query: 61 PSLDNAIEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHH 120
PSL AIEESRI IP+FSANYASSSFCLDELVHIIH YK G VLPVF+G DP+HVRH
Sbjct: 246 PSLVKAIEESRILIPIFSANYASSSFCLDELVHIIHCYKTRGCSVLPVFYGADPTHVRHQ 425
Query: 121 RGSYGEALAKHEERFQHNTDHMERLQKWKIALTQAANLSGDHRSPGYEYKLTGKI 175
GSYGE L KHE++FQ+N ++MERL+KWK+ALTQAAN SG H S GYEY+L I
Sbjct: 426 TGSYGEHLTKHEDKFQNNKENMERLKKWKMALTQAANFSGHHFSQGYEYELIENI 590
Score = 43.1 bits (100), Expect = 5e-04
Identities = 20/39 (51%), Positives = 27/39 (68%)
Frame = +2
Query: 196 QI*I*FYWRHSQIHLQQDQSCALTCCQLPCWI*VSNTTS 234
+I*+ Y +S+ H + DQSC T CQ+PCWI V +TTS
Sbjct: 560 RI*VRAY*EYSRTHFR*DQSCFSTRCQIPCWITVPSTTS 676
>BF651081 weakly similar to GP|18033111|g functional candidate resistance
protein KR1 {Glycine max}, partial (16%)
Length = 664
Score = 229 bits (585), Expect(2) = 1e-66
Identities = 117/147 (79%), Positives = 126/147 (85%), Gaps = 1/147 (0%)
Frame = +1
Query: 30 GFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIFIPVFSANYASSSFCLD 89
GFTGNLYKAL DKGIHTFID+ ELQRGDEITPSL AIEESRIFI VFS NYASSSFCLD
Sbjct: 67 GFTGNLYKALDDKGIHTFIDNHELQRGDEITPSLLKAIEESRIFIAVFSINYASSSFCLD 246
Query: 90 ELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEALAKHEERFQHNTDHMERLQKWK 149
ELVHIIH YK GRLVLPVFF V+P+ VRH +GSYGEALA+HE+RFQ++ MERLQ WK
Sbjct: 247 ELVHIIHCYKTKGRLVLPVFFAVEPTIVRHQKGSYGEALAEHEKRFQNDPKSMERLQGWK 426
Query: 150 IALTQAANLSGDHRS-PGYEYKLTGKI 175
AL+QAANLSG H S PGYEYKL GKI
Sbjct: 427 EALSQAANLSGYHDSPPGYEYKLIGKI 507
Score = 79.0 bits (193), Expect = 9e-15
Identities = 42/61 (68%), Positives = 45/61 (72%)
Frame = +3
Query: 197 I*I*FYWRHSQIHLQQDQSCALTCCQLPCWI*VSNTTSEIAS**GN**RGTHGRTLWDRR 256
I*I YW+ SQIHLQQDQS LTCC LPCWI VS TT+EIAS** * G +G LWD R
Sbjct: 480 I*IQAYWKDSQIHLQQDQSTTLTCCHLPCWIAVSXTTNEIAS**XI*SWGPYGWNLWDWR 659
Query: 257 L 257
L
Sbjct: 660 L 662
Score = 43.5 bits (101), Expect(2) = 1e-66
Identities = 19/23 (82%), Positives = 21/23 (90%)
Frame = +3
Query: 9 SISYVFKYQVFLSFRGSDTRYGF 31
SISY +KYQVFL+FRGSDTRY F
Sbjct: 3 SISYEYKYQVFLNFRGSDTRYXF 71
>AW688466 similar to GP|9965109|gb| resistance protein MG13 {Glycine max},
partial (31%)
Length = 668
Score = 249 bits (637), Expect = 3e-66
Identities = 135/235 (57%), Positives = 158/235 (66%)
Frame = +2
Query: 1 MQSSSSSSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEIT 60
MQS SSSSS+SY F + VF+SFRG+DTRYGFTGNLYKAL+DKGI TFIDD ELQRGDEIT
Sbjct: 41 MQSPSSSSSLSYGFSFDVFISFRGTDTRYGFTGNLYKALSDKGIRTFIDDKELQRGDEIT 220
Query: 61 PSLDNAIEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHH 120
PSL +IE+SRI I VFS +YASSSFCL ELVHII + G V+PVF+G +PS VRH
Sbjct: 221 PSLLKSIEDSRIAIIVFSKDYASSSFCLXELVHIIQCSNEKGTTVIPVFYGTEPSQVRHQ 400
Query: 121 RGSYGEALAKHEERFQHNTDHMERLQKWKIALTQAANLSGDHRSPGYEYKLTGKIAFNQT 180
SYGEALAKHEE FQ++ ++MERL KWK AL QAANLSG H
Sbjct: 401 NDSYGEALAKHEEGFQNSKENMERLLKWKKALNQAANLSGHH------------------ 526
Query: 181 PDLSSDCSQSLVEHEQI*I*FYWRHSQIHLQQDQSCALTCCQLPCWI*VSNTTSE 235
+ I* *FY ++ H QQD ++TCC+LP W +SN SE
Sbjct: 527 ----------FQSRK*I*T*FY*KNXYRHXQQD*PRSITCCRLPYWSQISNIRSE 661
>AW686165 weakly similar to GP|18033111|gb functional candidate resistance
protein KR1 {Glycine max}, partial (7%)
Length = 551
Score = 174 bits (441), Expect(2) = 9e-65
Identities = 83/109 (76%), Positives = 90/109 (82%)
Frame = +2
Query: 67 IEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGE 126
IEESRIFIPVFS NYASS FCLDELVHIIH YK GRLVLPVF+GVDP+ +RH GSYGE
Sbjct: 2 IEESRIFIPVFSINYASSKFCLDELVHIIHCYKTKGRLVLPVFYGVDPTQIRHQSGSYGE 181
Query: 127 ALAKHEERFQHNTDHMERLQKWKIALTQAANLSGDHRSPGYEYKLTGKI 175
L KHEE FQ+N + ERL +WK+ALTQAANLSG H SPGYEYK GKI
Sbjct: 182 HLTKHEESFQNNKKNKERLHQWKLALTQAANLSGYHYSPGYEYKFIGKI 328
Score = 92.4 bits (228), Expect(2) = 9e-65
Identities = 50/73 (68%), Positives = 55/73 (74%)
Frame = +1
Query: 197 I*I*FYWRHSQIHLQQDQSCALTCCQLPCWI*VSNTTSEIAS**GN**RGTHGRTLWDRR 256
I*I YW S+ HLQQDQSC TC Q+PCWI V TSEIAS** **RG +GRTLW R
Sbjct: 301 I*IQVYWEDSRRHLQQDQSCHFTCSQIPCWIRVPIRTSEIAS**RI**RGPYGRTLWYWR 480
Query: 257 LG*INTCKSNL*F 269
G*INTC+S+L*F
Sbjct: 481 FG*INTCQSDL*F 519
>TC84698 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (11%)
Length = 772
Score = 187 bits (476), Expect(3) = 2e-64
Identities = 89/113 (78%), Positives = 102/113 (89%)
Frame = +2
Query: 30 GFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIFIPVFSANYASSSFCLD 89
GFTGNLYKALTDKGIHTFIDD++LQRGDEITPSL AI+ESRIFIP+FSANYASSSFCL+
Sbjct: 302 GFTGNLYKALTDKGIHTFIDDNDLQRGDEITPSLVKAIDESRIFIPIFSANYASSSFCLN 481
Query: 90 ELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEALAKHEERFQHNTDHM 142
ELVHIIH YK GRLVLPVF GV+P++VRH GSYGEAL +HE+RFQ++ +M
Sbjct: 482 ELVHIIHCYKTKGRLVLPVFLGVEPTNVRHQTGSYGEALDEHEKRFQNDKSNM 640
Score = 52.4 bits (124), Expect(3) = 2e-64
Identities = 22/33 (66%), Positives = 28/33 (84%)
Frame = +3
Query: 143 ERLQKWKIALTQAANLSGDHRSPGYEYKLTGKI 175
ERLQ+WK+AL+QA+NLSG H PGY+Y+ GKI
Sbjct: 642 ERLQRWKVALSQASNLSGYHFCPGYQYEFIGKI 740
Score = 46.2 bits (108), Expect(3) = 2e-64
Identities = 22/31 (70%), Positives = 26/31 (82%)
Frame = +1
Query: 1 MQSSSSSSSISYVFKYQVFLSFRGSDTRYGF 31
+QS SSSSS S+ F YQVFLSFRG+DTR+ F
Sbjct: 214 IQSPSSSSSFSFGFTYQVFLSFRGTDTRHWF 306
>BF636566 weakly similar to GP|9965103|gb| resistance protein LM6 {Glycine
max}, partial (20%)
Length = 663
Score = 234 bits (597), Expect(2) = 5e-64
Identities = 136/209 (65%), Positives = 154/209 (73%)
Frame = +2
Query: 262 TCKSNL*FYW*SI*RIMFSS*CEREFS*K*LETSPREAPFENNWIGN*VRPC**GNSNH* 321
TCKSNL*F SI*R MFSS* REF+ *LETSP A F+N+ I +*VR C**GNS +
Sbjct: 2 TCKSNL*FCCRSI*RCMFSS*S*REFNS**LETSPEGASFKNS*IEH*VRRC**GNSTYK 181
Query: 322 GKAM*KEDSFDSR*C*QYESTACFGWRT*LVRPWKQSHHYHSRQTLTK*SWDKKYTCSRR 381
GK * EDSFDSR*C*Q + WRT*LV WK SHHYHSR TLT WD+K C +R
Sbjct: 182 GKTK*NEDSFDSR*C*QTGTARGISWRT*LVWSWK*SHHYHSR*TLTNMPWDRKNICGKR 361
Query: 382 VEWD*SS*IVKVDGF*K*QSSFRLRRHFKPCSCIFFWPSISYRSSGFKLVWKEHRKMEEY 441
D S *IV+VDGF*K SSF L R FKPCSCI F PS+S R+SGF LVWKEH +M +
Sbjct: 362 AT*DRSF*IVEVDGF*KW*SSFEL*RCFKPCSCICFRPSVSSRNSGF*LVWKEHGRMAMH 541
Query: 442 IRWV*QDSQ*RDPKDT*SKL*CFGRRGAE 470
IRWV*+DSQ*RDPKDT*SKL*CF ++ ++
Sbjct: 542 IRWV*KDSQ*RDPKDT*SKL*CFWKKSSK 628
Score = 30.0 bits (66), Expect(2) = 5e-64
Identities = 12/18 (66%), Positives = 14/18 (77%)
Frame = +3
Query: 464 FGRRGAECLSGHCLLLQR 481
FGRR A+CLS +CL QR
Sbjct: 609 FGRRAAKCLSRYCLXFQR 662
>TC77728 similar to GP|12056928|gb|AAG48132.1 putative resistance protein
{Glycine max}, partial (12%)
Length = 765
Score = 238 bits (607), Expect = 8e-63
Identities = 119/160 (74%), Positives = 133/160 (82%), Gaps = 1/160 (0%)
Frame = +1
Query: 17 QVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIFIPV 76
+VFLSFRGSDTR FTGNLYKAL DKGI TFIDD++L+RGDEITPSL AIEESRIFIP+
Sbjct: 91 RVFLSFRGSDTRNKFTGNLYKALVDKGIRTFIDDNDLERGDEITPSLVKAIEESRIFIPI 270
Query: 77 FSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEALAKHEERFQ 136
FSANYASSSFCLDELVHIIH YK LV PVF+ V+P+H+R+ G YGE L KHEERFQ
Sbjct: 271 FSANYASSSFCLDELVHIIHCYKTKSCLVFPVFYDVEPTHIRNQSGIYGEHLTKHEERFQ 450
Query: 137 HNTDHMERLQKWKIALTQAANLSGDHRSP-GYEYKLTGKI 175
+N +MERL++WKIAL QAANLSG H SP GYEYK KI
Sbjct: 451 NNEKNMERLRQWKIALIQAANLSGYHYSPHGYEYKFIEKI 570
Score = 79.0 bits (193), Expect = 9e-15
Identities = 46/73 (63%), Positives = 49/73 (67%)
Frame = +3
Query: 197 I*I*FYWRHSQIHLQQDQSCALTCCQLPCWI*VSNTTSEIAS**GN**RGTHGRTLWDRR 256
I*I Y S+ HLQQ QSC C Q+PCWI V N SEIAS* G * G HGRT W R
Sbjct: 543 I*IQVY*EDSRRHLQQYQSCFFKCSQIPCWITVPNRRSEIAS*HGL*G*GPHGRTFWYWR 722
Query: 257 LG*INTCKSNL*F 269
G*INTCKS+L*F
Sbjct: 723 HG*INTCKSSL*F 761
>AW559878 weakly similar to GP|18033111|g functional candidate resistance
protein KR1 {Glycine max}, partial (12%)
Length = 586
Score = 237 bits (605), Expect = 1e-62
Identities = 120/176 (68%), Positives = 139/176 (78%), Gaps = 1/176 (0%)
Frame = +2
Query: 1 MQSSSSSSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEIT 60
+ SSSSSSS+S F Y VF+SFRG DTR GFTG+LYKAL DKGI TFIDD ELQRGDEIT
Sbjct: 56 LPSSSSSSSLSNDFIYDVFISFRGIDTRSGFTGHLYKALCDKGIRTFIDDKELQRGDEIT 235
Query: 61 PSLDNAIEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHH 120
PSL +IE S+I I VFS NYA+SSFCLDELVHII+ +K+ GRLVLPVF+GV+PSHVRH
Sbjct: 236 PSLLKSIEHSKIAIIVFSENYATSSFCLDELVHIINYFKEKGRLVLPVFYGVEPSHVRHQ 415
Query: 121 RGSYGEALAKHEERFQHNTDHMERLQKWKIALTQAANLSGDH-RSPGYEYKLTGKI 175
YGEAL + EE FQ+N ++M+RLQKWKIAL Q NLSG H + YEY+ KI
Sbjct: 416 NNKYGEALTEFEEMFQNNKENMDRLQKWKIALNQVGNLSGXHFKKDAYEYEFIKKI 583
>TC77733 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (12%)
Length = 632
Score = 236 bits (602), Expect = 3e-62
Identities = 115/149 (77%), Positives = 128/149 (85%)
Frame = +3
Query: 17 QVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIFIPV 76
+VFLSFRGSDTR FTGNLYKAL DKGI TFIDD++L+RGDEITPSL AIEESRIFIP+
Sbjct: 51 RVFLSFRGSDTRNNFTGNLYKALVDKGIRTFIDDNDLERGDEITPSLVKAIEESRIFIPI 230
Query: 77 FSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEALAKHEERFQ 136
FSANYASSSFCLDELVHIIH YK LVLPVF+ V+P+H+RH GSYGE L KHEERFQ
Sbjct: 231 FSANYASSSFCLDELVHIIHCYKTKSCLVLPVFYDVEPTHIRHQSGSYGEHLTKHEERFQ 410
Query: 137 HNTDHMERLQKWKIALTQAANLSGDHRSP 165
+N +MERL++WKIAL QAANLSG H SP
Sbjct: 411 NNEKNMERLRQWKIALIQAANLSGYHYSP 497
Score = 32.7 bits (73), Expect = 0.70
Identities = 16/21 (76%), Positives = 17/21 (80%)
Frame = +2
Query: 567 GMAFKKMTKLKTLIIENGHFS 587
GMA KKMT LKTLIIE +FS
Sbjct: 569 GMACKKMTNLKTLIIEYANFS 631
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.355 0.155 0.549
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,712,632
Number of Sequences: 36976
Number of extensions: 532642
Number of successful extensions: 6696
Number of sequences better than 10.0: 187
Number of HSP's better than 10.0 without gapping: 2125
Number of HSP's successfully gapped in prelim test: 304
Number of HSP's that attempted gapping in prelim test: 4330
Number of HSP's gapped (non-prelim): 2693
length of query: 984
length of database: 9,014,727
effective HSP length: 105
effective length of query: 879
effective length of database: 5,132,247
effective search space: 4511245113
effective search space used: 4511245113
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC123574.2


