

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123574.1 + phase: 0 /pseudo
(527 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
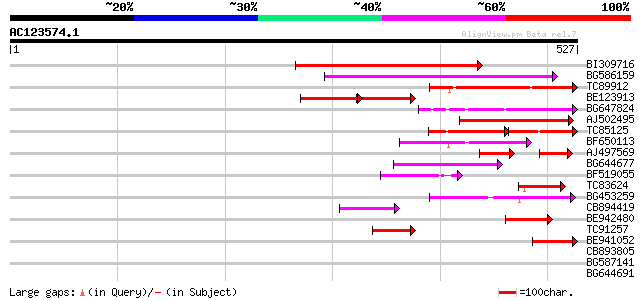
Score E
Sequences producing significant alignments: (bits) Value
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 194 5e-50
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 161 7e-40
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 99 4e-21
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 62 3e-20
BG647824 weakly similar to PIR|G96722|G96 hypothetical protein F... 92 5e-19
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 92 6e-19
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 57 2e-18
BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vu... 79 4e-15
AJ497569 weakly similar to PIR|T04833|T04 hypothetical protein F... 49 4e-12
BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T... 56 3e-08
BF519055 similar to GP|21592754|gb| unknown {Arabidopsis thalian... 52 4e-07
TC83624 homologue to PIR|G84581|G84581 copia-like retroelement p... 50 2e-06
BG453259 homologue to GP|21434|emb|CA ORF4 {Solanum tuberosum}, ... 50 3e-06
CB894419 weakly similar to GP|10177935|db copia-type polyprotein... 46 3e-05
BE942480 46 3e-05
TC91257 similar to GP|21434|emb|CAA36616.1|| ORF4 {Solanum tuber... 44 1e-04
BE941052 weakly similar to PIR|B85188|B85 retrotransposon like p... 44 2e-04
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 39 0.005
BG587141 similar to PIR|H86461|H86 hypothetical protein AAF32440... 39 0.006
BG644691 similar to GP|21434|emb|CA ORF4 {Solanum tuberosum}, pa... 38 0.011
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 194 bits (494), Expect = 5e-50
Identities = 99/174 (56%), Positives = 125/174 (70%)
Frame = +2
Query: 266 SEFNNIKSVLRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPI 325
SE ++K L FKIKDLG L+YFLGL+VA SKQGI L QRKY L+LL DSG K
Sbjct: 218 SEIQHVKCFLIDRFKIKDLGSLRYFLGLEVARSKQGILLNQRKYTLELLEDSGNLAVKST 397
Query: 326 STPSDPSVKLHRDSSPPYTDISAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHH 385
TP D S+KLH SP Y D + YRRL+G+L+YL TTRPDI+F QQLSQF++K QVH+
Sbjct: 398 LTPYDISLKLHNSDSPLYNDETQYRRLIGKLIYLTTTRPDISFAVQQLSQFVSKPQQVHY 577
Query: 386 SAAMRVLKYLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLG 439
AA+RVL+YLKT+ KGLF+ S L++ F+D+DWA C +R+S++G FLG
Sbjct: 578 QAAIRVLQYLKTAPAKGLFYSATSNLKLSSFADSDWATCPTTRKSVTGYWVFLG 739
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 161 bits (407), Expect = 7e-40
Identities = 78/217 (35%), Positives = 129/217 (58%)
Frame = +1
Query: 293 LQVAHSKQGISLCQRKYCLDLLADSGLTHSKPISTPSDPSVKLHRDSSPPYTDISAYRRL 352
++V +++GI +CQRKY DLL G+ S P P KL +D + D + Y+++
Sbjct: 1 VEVIQNEEGIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIKDENGVKVDATKYKQI 180
Query: 353 VGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHHSAAMRVLKYLKTSLGKGLFFPRDSALQ 412
VG L+YL TRPD+ +V +S+F+ T++H A RVL+YL ++ G+ + R+ + +
Sbjct: 181 VGCLMYLAATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMYKRNGSEK 360
Query: 413 ILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKKQLIVSRSSSKEEYRALAAATCEL 472
+ ++D+D+AG LD R+S SG F L +SW +KKQ +V+ S++K E+ A A C+
Sbjct: 361 LEAYTDSDYAGDLDDRKSTSGYVFMLSSGAVSWSSKKQPVVTLSTTKAEFIAAAFCACQS 540
Query: 473 QWLLYLLQDLQLQTSKLHVLYCDNQSALHIAANPIFH 509
W+ +L+ L S +YCDN S + ++ NP+ H
Sbjct: 541 VWMRRVLEKLGYTQSGSITMYCDNNSTIKLSKNPVLH 651
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 99.0 bits (245), Expect = 4e-21
Identities = 52/141 (36%), Positives = 90/141 (62%), Gaps = 4/141 (2%)
Frame = +1
Query: 391 VLKYLKTSLGKGLFFPR----DSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISWR 446
VLKYL SL L + + + AL+ G+ DAD+AG +D+R+S+SG F L + ISW+
Sbjct: 16 VLKYLNESLKSSLKYTKAAQEEDALE--GYVDADYAGNVDTRKSLSGFVFTLYGTTISWK 189
Query: 447 TKKQLIVSRSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAANP 506
+Q +V+ S+++ EY A + WL ++ +L + T + ++CD+QSA+H+A +
Sbjct: 190 ANQQSVVTLSTTQAEYIAFVEGVKDAIWLKGMIGELGI-TQEYVKIHCDSQSAIHLANHQ 366
Query: 507 IFHERTKHLEIDCHLVQDKLQ 527
++HERTKH++I H ++D ++
Sbjct: 367 VYHERTKHIDIRLHFIRDMIE 429
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 62.0 bits (149), Expect(2) = 3e-20
Identities = 30/58 (51%), Positives = 41/58 (69%)
Frame = +1
Query: 271 IKSVLRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPISTP 328
+K++L F+IKDLG LKYFLG++VA K+G S+ QRKY LDLL ++ + K I P
Sbjct: 181 LKNLLAEEFEIKDLGNLKYFLGMEVARWKKGSSISQRKYVLDLLKETRMIGCKTIRDP 354
Score = 54.7 bits (130), Expect(2) = 3e-20
Identities = 27/55 (49%), Positives = 36/55 (65%)
Frame = +2
Query: 323 KPISTPSDPSVKLHRDSSPPYTDISAYRRLVGRLLYLNTTRPDITFVTQQLSQFL 377
KP TP D +VKL + D Y+RLVG+L+YL+ TRPDI+FV +SQF+
Sbjct: 338 KPSETPMDATVKLGTLDNGTLVDKGRYQRLVGKLIYLSHTRPDISFVVCTMSQFM 502
>BG647824 weakly similar to PIR|G96722|G96 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (5%)
Length = 721
Score = 92.0 bits (227), Expect = 5e-19
Identities = 62/148 (41%), Positives = 85/148 (56%), Gaps = 1/148 (0%)
Frame = -3
Query: 381 TQVHHSAAMRVLKYLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGK 440
T H AA+++L YLK S + +FFP S +QI F D+D + ++ Q
Sbjct: 599 TAQHPQAAIQIL-YLKISPSQ*IFFP--S*IQIKAFCDSD*ID--QAA*TLENQSVIFAS 435
Query: 441 SLISWRTKKQLIVSRSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSAL 500
S + L R + K YR++ + CE++WL YLL DL+ K +LYCDNQSA
Sbjct: 434 S*ATHSYAGNLKKKRYNFKI-YRSI*STICEIKWLTYLLNDLKFTFIKPAMLYCDNQSAA 258
Query: 501 -HIAANPIFHERTKHLEIDCHLVQDKLQ 527
HIAAN F ERTKH+E+DCH+V+ KLQ
Sbjct: 257 RHIAANSSFLERTKHIELDCHIVRVKLQ 174
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 91.7 bits (226), Expect = 6e-19
Identities = 40/106 (37%), Positives = 69/106 (64%)
Frame = +2
Query: 419 ADWAGCLDSRRSISGQCFFLGKSLISWRTKKQLIVSRSSSKEEYRALAAATCELQWLLYL 478
+DWAG ++R+S SG F LG ISW +KKQ +V+ S+++ EY A + + WL +
Sbjct: 2 SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQPVVAFSTAEAEYIASTSCATQTVWLRRI 181
Query: 479 LQDLQLQTSKLHVLYCDNQSALHIAANPIFHERTKHLEIDCHLVQD 524
L+ + + + +YCDN+SA+ ++ NP+FH R+KH++I H +++
Sbjct: 182 LEVMHHEQNTPTKIYCDNKSAIALSKNPVFHGRSKHIDIQFHKIRE 319
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 57.4 bits (137), Expect(2) = 2e-18
Identities = 28/64 (43%), Positives = 42/64 (64%)
Frame = +3
Query: 464 ALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAANPIFHERTKHLEIDCHLVQ 523
+L A E W+ L+++L + ++ V YCD+QSALHIA NP FH RTKH+ I H V+
Sbjct: 228 SLPQACKEAIWMQRLMEELGHKQEQITV-YCDSQSALHIARNPAFHSRTKHIGIQYHFVR 404
Query: 524 DKLQ 527
+ ++
Sbjct: 405 EVVE 416
Score = 53.1 bits (126), Expect(2) = 2e-18
Identities = 27/75 (36%), Positives = 46/75 (61%)
Frame = +1
Query: 390 RVLKYLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKK 449
R+++Y+K + G + F S L + G+ D+D+AG D R+S +G F L +SW +K
Sbjct: 7 RIMRYIKGTSGVAVCFG-GSELTVRGYVDSDFAGDHDKRKSTTGYVFTLAGGAVSWLSKL 183
Query: 450 QLIVSRSSSKEEYRA 464
Q +V+ S+++ EY A
Sbjct: 184 QTVVALSTTEAEYMA 228
>BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vulgaris},
partial (13%)
Length = 494
Score = 79.0 bits (193), Expect = 4e-15
Identities = 45/126 (35%), Positives = 72/126 (56%), Gaps = 3/126 (2%)
Frame = +1
Query: 363 RPDITFVTQQLSQFLAKRTQVHHSAAMRVLKYLKTSLGKGLFFP---RDSALQILGFSDA 419
RPDI + +S+F+ + H AA R+L+Y++ ++ GL FP + +++ +SD+
Sbjct: 121 RPDICYSVSVISKFMHDPRKPHLIAANRILRYVRGTMEYGLLFPYGAKSEVYELICYSDS 300
Query: 420 DWAGCLDSRRSISGQCFFLGKSLISWRTKKQLIVSRSSSKEEYRALAAATCELQWLLYLL 479
DW G RRS SG F + ISW TKKQ I + SS + EY A AT + WL ++
Sbjct: 301 DWCG---DRRSTSGYVFKFNDAAISWCTKKQPITALSSYEAEYIAGTFATFQALWLDSVI 471
Query: 480 QDLQLQ 485
++L+ +
Sbjct: 472 KELKCE 489
>AJ497569 weakly similar to PIR|T04833|T04 hypothetical protein F21P8.50 -
Arabidopsis thaliana, partial (4%)
Length = 723
Score = 48.9 bits (115), Expect(2) = 4e-12
Identities = 22/31 (70%), Positives = 25/31 (79%)
Frame = +2
Query: 493 YCDNQSALHIAANPIFHERTKHLEIDCHLVQ 523
YCDN SALHIAAN +FHERT H E D ++VQ
Sbjct: 290 YCDNISALHIAANMVFHERT*HRETDPYIVQ 382
Score = 40.0 bits (92), Expect(2) = 4e-12
Identities = 21/33 (63%), Positives = 25/33 (75%)
Frame = +3
Query: 437 FLGKSLISWRTKKQLIVSRSSSKEEYRALAAAT 469
FL SLISW++KKQ +VSRS S+ RALA AT
Sbjct: 126 FLSSSLISWKSKKQCVVSRSFSEA**RALANAT 224
>BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T15F17.l
{Arabidopsis thaliana}, partial (3%)
Length = 539
Score = 56.2 bits (134), Expect = 3e-08
Identities = 31/102 (30%), Positives = 54/102 (52%)
Frame = -3
Query: 357 LYLNTTRPDITFVTQQLSQFLAKRTQVHHSAAMRVLKYLKTSLGKGLFFPRDSALQILGF 416
+ L P+ITF LS++ + T H + + KYLK + GLF+ +D + ++G+
Sbjct: 531 ILLTLQGPNITFSINLLSRYSSAPTMRH*NGIKHICKYLKGIIDMGLFYSKDCSPDLIGY 352
Query: 417 SDADWAGCLDSRRSISGQCFFLGKSLISWRTKKQLIVSRSSS 458
+A + RS +G F G ++ISWR+ K ++ SS+
Sbjct: 351 VNA*YLSDPHKARS*TGYIFTCGNTVISWRSTK*STIATSSN 226
>BF519055 similar to GP|21592754|gb| unknown {Arabidopsis thaliana}, partial
(30%)
Length = 675
Score = 52.4 bits (124), Expect = 4e-07
Identities = 30/77 (38%), Positives = 44/77 (56%)
Frame = -3
Query: 345 DISAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHHSAAMRVLKYLKTSLGKGLF 404
D +VG LLY+ T PD++F + SQF+ K TQ++ +V+++ K ++ K LF
Sbjct: 646 DSKLVHSIVGALLYITVTCPDLSFSINKPSQFMHKPTQINFQQLKKVMRHPKLTI-KKLF 470
Query: 405 FPRDSALQILGFSDADW 421
LQI FSDADW
Sbjct: 469 -----DLQIYAFSDADW 434
>TC83624 homologue to PIR|G84581|G84581 copia-like retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(1%)
Length = 831
Score = 50.1 bits (118), Expect = 2e-06
Identities = 23/48 (47%), Positives = 34/48 (69%), Gaps = 5/48 (10%)
Frame = +2
Query: 474 WLLY-----LLQDLQLQTSKLHVLYCDNQSALHIAANPIFHERTKHLE 516
W+LY L++LQ+Q ++L ++YC NQ L+IA N ++HERTKH E
Sbjct: 413 WILYNGCYTFLRNLQVQCTRLLLIYCVNQITLYIAKNQVYHERTKH*E 556
>BG453259 homologue to GP|21434|emb|CA ORF4 {Solanum tuberosum}, partial (5%)
Length = 657
Score = 49.7 bits (117), Expect = 3e-06
Identities = 39/138 (28%), Positives = 67/138 (48%), Gaps = 2/138 (1%)
Frame = -2
Query: 391 VLKYLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKKQ 450
+++ L +S + + F R+ L + + + AG + R S SG FLG +++ KQ
Sbjct: 566 LIEGLXSSGTQMIVFKRE*KLSMEVYXNTVCAGWIVDRGSTSGY*MFLGGNMVE*---KQ 396
Query: 451 LIVSRSSSKEEYRALAAATCEL--QWLLYLLQDLQLQTSKLHVLYCDNQSALHIAANPIF 508
+V+R + + + + + L L DL + L+ +N IA NP+
Sbjct: 395 NVVAR*VQRHNFELCSQGL*RVMDEELKIKLDDLIINYKDPMTLF*NNNFVSRIAHNPVQ 216
Query: 509 HERTKHLEIDCHLVQDKL 526
H RTKH+EID H + +KL
Sbjct: 215 HYRTKHIEIDQHFIIEKL 162
>CB894419 weakly similar to GP|10177935|db copia-type polyprotein
{Arabidopsis thaliana}, partial (2%)
Length = 170
Score = 46.2 bits (108), Expect = 3e-05
Identities = 23/56 (41%), Positives = 32/56 (57%)
Frame = +3
Query: 307 RKYCLDLLADSGLTHSKPISTPSDPSVKLHRDSSPPYTDISAYRRLVGRLLYLNTT 362
+K D+L + HSKPISTP + +KL R+S D + Y+ L+G L YL T
Sbjct: 3 KKCASDILKKFKMEHSKPISTPVEEKLKLTRESDGKRVDSTHYKSLIGSLRYLTAT 170
>BE942480
Length = 396
Score = 46.2 bits (108), Expect = 3e-05
Identities = 19/43 (44%), Positives = 32/43 (74%)
Frame = -2
Query: 462 YRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAA 504
YRA+++ CE++WL Y++ L++Q+ K + Y DNQ+A HIA+
Sbjct: 317 YRAMSSIVCEIEWLTYIVDVLKVQSIKPTLPYYDNQAARHIAS 189
>TC91257 similar to GP|21434|emb|CAA36616.1|| ORF4 {Solanum tuberosum},
partial (8%)
Length = 854
Score = 44.3 bits (103), Expect = 1e-04
Identities = 20/40 (50%), Positives = 26/40 (65%)
Frame = +3
Query: 338 DSSPPYTDISAYRRLVGRLLYLNTTRPDITFVTQQLSQFL 377
D ++D YRRLVG+L YL TRPDI++ +SQFL
Sbjct: 729 DQGETFSDPGRYRRLVGKLNYLTMTRPDISYAVSVVSQFL 848
>BE941052 weakly similar to PIR|B85188|B85 retrotransposon like protein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 480
Score = 43.9 bits (102), Expect = 2e-04
Identities = 18/41 (43%), Positives = 27/41 (64%)
Frame = +2
Query: 487 SKLHVLYCDNQSALHIAANPIFHERTKHLEIDCHLVQDKLQ 527
S+ +L CD SA ++ NP++H R KH+ ID H V+D +Q
Sbjct: 8 SQSGLLRCDYLSATYLTHNPVYHSRMKHISIDIHFVRDLVQ 130
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 38.9 bits (89), Expect = 0.005
Identities = 14/38 (36%), Positives = 25/38 (64%)
Frame = +3
Query: 268 FNNIKSVLRASFKIKDLGQLKYFLGLQVAHSKQGISLC 305
F K ++ F + DLG++ YFLG++V +++GI +C
Sbjct: 663 FEEFKKSMKKEFNMSDLGKMHYFLGVEVTQNEKGIYIC 776
>BG587141 similar to PIR|H86461|H86 hypothetical protein AAF32440.1
[imported] - Arabidopsis thaliana, partial (20%)
Length = 731
Score = 38.5 bits (88), Expect = 0.006
Identities = 17/57 (29%), Positives = 30/57 (51%)
Frame = +3
Query: 468 ATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAANPIFHERTKHLEIDCHLVQD 524
A + WL LL ++ + + V+ DNQS + + NP+FH R H+ H +++
Sbjct: 126 AARQAMWLQDLLSEVTWEPCEEVVIRIDNQSVIALTRNPVFHGRGNHIHKRYHFIRE 296
>BG644691 similar to GP|21434|emb|CA ORF4 {Solanum tuberosum}, partial (5%)
Length = 753
Score = 37.7 bits (86), Expect = 0.011
Identities = 20/37 (54%), Positives = 24/37 (64%), Gaps = 1/37 (2%)
Frame = +1
Query: 488 KLHVLYC-DNQSALHIAANPIFHERTKHLEIDCHLVQ 523
K + +C DN + IA NPI H+RTKH EID HL Q
Sbjct: 421 KCRLPFCYDNIIPISIAHNPI*HDRTKHTEIDRHLHQ 531
Score = 26.9 bits (58), Expect(2) = 0.62
Identities = 14/22 (63%), Positives = 15/22 (67%)
Frame = +3
Query: 411 LQILGFSDADWAGCLDSRRSIS 432
LQ GF+DA G LD RRSIS
Sbjct: 42 LQTEGFTDAH*NGSLDDRRSIS 107
Score = 23.5 bits (49), Expect(2) = 0.62
Identities = 12/37 (32%), Positives = 19/37 (50%)
Frame = +2
Query: 445 WRTKKQLIVSRSSSKEEYRALAAATCELQWLLYLLQD 481
WR+ K +V+ SS +YRA+ LL++ D
Sbjct: 143 WRS*KHNVVALSSV*ADYRAMNRVFMSFFSLLFIFDD 253
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.355 0.156 0.561
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,619,676
Number of Sequences: 36976
Number of extensions: 360769
Number of successful extensions: 4689
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 2672
Number of HSP's successfully gapped in prelim test: 226
Number of HSP's that attempted gapping in prelim test: 1938
Number of HSP's gapped (non-prelim): 3043
length of query: 527
length of database: 9,014,727
effective HSP length: 101
effective length of query: 426
effective length of database: 5,280,151
effective search space: 2249344326
effective search space used: 2249344326
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC123574.1


