

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
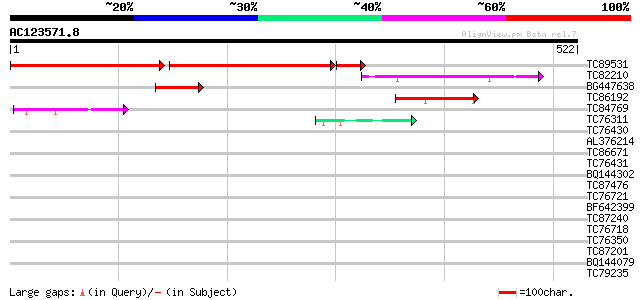
Query= AC123571.8 - phase: 0
(522 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC89531 similar to GP|19034049|gb|AAL83681.1 putative extensin {... 327 0.0
TC82210 weakly similar to GP|15450527|gb|AAK96556.1 AT5g21160/T1... 100 1e-21
BG447638 92 5e-19
TC86192 weakly similar to GP|20161211|dbj|BAB90138. RNA-binding ... 55 7e-08
TC84769 similar to PIR|E96665|E96665 protein F22C12.16 [imported... 55 9e-08
TC76311 homologue to GP|3204132|emb|CAA07235.1 extensin {Cicer a... 42 6e-04
TC76430 similar to PIR|T07623|T07623 extensin homolog HRGP2 - so... 40 0.002
AL376214 homologue to GP|3204132|emb| extensin {Cicer arietinum}... 39 0.004
TC86671 similar to PIR|T06782|T06782 extensin - soybean, partial... 39 0.005
TC76431 similar to GP|8132441|gb|AAF73291.1| extensin {Pisum sat... 39 0.005
BQ144302 similar to GP|6815109|dbj| hypothetical protein {Oryza ... 38 0.011
TC87476 homologue to PIR|T11622|T11622 extensin class 1 precurso... 37 0.014
TC76721 homologue to GP|8132441|gb|AAF73291.1| extensin {Pisum s... 37 0.024
BF642399 similar to PIR|T10863|T10 extensin precursor - kidney b... 37 0.024
TC87240 similar to GP|19571126|dbj|BAB86550. putative 5-3 exorib... 37 0.024
TC76718 homologue to GP|8132441|gb|AAF73291.1| extensin {Pisum s... 37 0.024
TC76350 similar to GP|8132441|gb|AAF73291.1| extensin {Pisum sat... 36 0.032
TC87201 similar to PIR|F86388|F86388 hypothetical protein AAG292... 36 0.041
BQ144079 similar to GP|6523547|emb| hydroxyproline-rich glycopro... 36 0.041
TC79235 weakly similar to GP|7297741|gb|AAF52992.1| CG17108 gene... 36 0.041
>TC89531 similar to GP|19034049|gb|AAL83681.1 putative extensin {Oryza
sativa (japonica cultivar-group)}, partial (8%)
Length = 1184
Score = 327 bits (839), Expect(3) = 0.0
Identities = 151/153 (98%), Positives = 152/153 (98%)
Frame = +3
Query: 148 VGDNVNVNNMAPTRQKSIKHNSSNASSNGGHTQQSAPQVSIAATGSHTSSSRDHTQSPRD 207
VGDNVNVNNMAPTRQKSIKHNSSNASSNGGHTQQS PQV+IAATGSHTSSSRDHTQSPRD
Sbjct: 642 VGDNVNVNNMAPTRQKSIKHNSSNASSNGGHTQQSEPQVTIAATGSHTSSSRDHTQSPRD 821
Query: 208 HTQSPRDHAQSPRDHTQRSGFVPSDHPQQRNSFRHRNGGPHQRGDGSHHHHNYGNRRDQD 267
HTQSPRDHAQSPRDHTQRSGFVPSDHPQQRNSFRHRNGGPHQRGDGSHHHHNYGNRRDQD
Sbjct: 822 HTQSPRDHAQSPRDHTQRSGFVPSDHPQQRNSFRHRNGGPHQRGDGSHHHHNYGNRRDQD 1001
Query: 268 WNSRRNYNGRDMHVPPRVSPRIIRPSLPPNSAP 300
WNSRRNYNGRDMHVPPRVSPRIIRPSLPPNSAP
Sbjct: 1002WNSRRNYNGRDMHVPPRVSPRIIRPSLPPNSAP 1100
Score = 288 bits (738), Expect(3) = 0.0
Identities = 142/142 (100%), Positives = 142/142 (100%)
Frame = +1
Query: 1 MEMIGNHSSNHSPDNLQSRRLPPWNQVVRGESESIAAVPAVSLSEESFPIVTAPVDDSTS 60
MEMIGNHSSNHSPDNLQSRRLPPWNQVVRGESESIAAVPAVSLSEESFPIVTAPVDDSTS
Sbjct: 199 MEMIGNHSSNHSPDNLQSRRLPPWNQVVRGESESIAAVPAVSLSEESFPIVTAPVDDSTS 378
Query: 61 AEVISDNADNGGERNGGTGKRPAWNRSSGNGGVSEVQPVMDAHSWPALSDSARGSTKSES 120
AEVISDNADNGGERNGGTGKRPAWNRSSGNGGVSEVQPVMDAHSWPALSDSARGSTKSES
Sbjct: 379 AEVISDNADNGGERNGGTGKRPAWNRSSGNGGVSEVQPVMDAHSWPALSDSARGSTKSES 558
Query: 121 SKGLLDGSSVSPWQGMESTPSS 142
SKGLLDGSSVSPWQGMESTPSS
Sbjct: 559 SKGLLDGSSVSPWQGMESTPSS 624
Score = 60.8 bits (146), Expect(3) = 0.0
Identities = 26/26 (100%), Positives = 26/26 (100%)
Frame = +1
Query: 302 IHPPPLRPFGGHMGFHELAAPVVLFA 327
IHPPPLRPFGGHMGFHELAAPVVLFA
Sbjct: 1105 IHPPPLRPFGGHMGFHELAAPVVLFA 1182
>TC82210 weakly similar to GP|15450527|gb|AAK96556.1 AT5g21160/T10F18_190
{Arabidopsis thaliana}, partial (24%)
Length = 1436
Score = 100 bits (250), Expect = 1e-21
Identities = 64/182 (35%), Positives = 90/182 (49%), Gaps = 15/182 (8%)
Frame = +2
Query: 325 LFAGPPPPIDSLRGVPFVPPMPLYYAGPDPQ---LHSKIVNQIDYYFSNENLVKDIFLRK 381
L GPPP R PP+ P P+ L + I+ QI+YYFS+ENL D +L
Sbjct: 11 LLRGPPP-----RHFAPYPPVNSAPQSPTPETQSLRASILKQIEYYFSDENLHNDRYLIG 175
Query: 382 NMDAQGWVPITLIAGFKKVMDLTDNIQLIIDAIRTSSVVEVQGDKIRRQNDWEKWIMPS- 440
MD QGWVPI+ +A FK+V ++ +I I+D ++ S VEVQ DKIR++N+W KWI S
Sbjct: 176 LMDDQGWVPISTVADFKRVKRMSTDIPFIVDVLQNSDNVEVQDDKIRKRNNWSKWIQTSS 355
Query: 441 -----------PVQFPNVTSPEVLNQDMLAEKMRNIALETTIYDGAGGPVVLPDNSEHTP 489
Q T+ N D + +K + + E T+ D A N +
Sbjct: 356 GNSGSSVAQVQQDQHVESTANSCQNSDTVVDKTKE-SSEATLNDSAHDSTSTEQNQSNKD 532
Query: 490 AF 491
F
Sbjct: 533 TF 538
Score = 33.9 bits (76), Expect = 0.16
Identities = 36/126 (28%), Positives = 50/126 (39%), Gaps = 28/126 (22%)
Frame = +2
Query: 64 ISDNADNGGERNGGTGKRPAWNR----SSGNGGVSEVQPVMDAH---------------- 103
+ N+DN ++ KR W++ SSGN G S Q D H
Sbjct: 269 VLQNSDNVEVQDDKIRKRNNWSKWIQTSSGNSGSSVAQVQQDQHVESTANSCQNSDTVVD 448
Query: 104 -----SWPALSDSARGSTKSE---SSKGLLDGSSVSPWQGMESTPSSSMQRQVGDNVNVN 155
S L+DSA ST +E S+K + S V+ Q S PS ++ V +N
Sbjct: 449 KTKESSEATLNDSAHDSTSTEQNQSNKDTFEVSDVNQKQDTNSHPSKNISHAV-----MN 613
Query: 156 NMAPTR 161
A TR
Sbjct: 614 KNATTR 631
>BG447638
Length = 394
Score = 92.0 bits (227), Expect = 5e-19
Identities = 44/44 (100%), Positives = 44/44 (100%)
Frame = +1
Query: 135 GMESTPSSSMQRQVGDNVNVNNMAPTRQKSIKHNSSNASSNGGH 178
GMESTPSSSMQRQVGDNVNVNNMAPTRQKSIKHNSSNASSNGGH
Sbjct: 262 GMESTPSSSMQRQVGDNVNVNNMAPTRQKSIKHNSSNASSNGGH 393
>TC86192 weakly similar to GP|20161211|dbj|BAB90138. RNA-binding
protein-like {Oryza sativa (japonica cultivar-group)},
partial (34%)
Length = 1617
Score = 55.1 bits (131), Expect = 7e-08
Identities = 31/80 (38%), Positives = 50/80 (61%), Gaps = 4/80 (5%)
Frame = +2
Query: 356 LHSKIVNQIDYYFSNENLVKDIFLRK--NMDAQGWVPITLIAGFKKVMDLTDNIQLIIDA 413
L KI+ Q++YYFS+ENL D ++ + +G+VPI IA F+K+ LT + I A
Sbjct: 344 LKLKIIKQVEYYFSDENLPTDKYMLSLVKRNKEGFVPIPAIASFRKMKRLTRDHLFIAAA 523
Query: 414 IRTSSVVEVQGD--KIRRQN 431
++ SS++ V GD +++R N
Sbjct: 524 LKESSLLVVSGDGKRVKRLN 583
>TC84769 similar to PIR|E96665|E96665 protein F22C12.16 [imported] -
Arabidopsis thaliana, partial (4%)
Length = 723
Score = 54.7 bits (130), Expect = 9e-08
Identities = 36/114 (31%), Positives = 54/114 (46%), Gaps = 8/114 (7%)
Frame = +3
Query: 4 IGNHSSNHSPD---NLQSRRLPPWNQVVRGESESIAAVPA-----VSLSEESFPIVTAPV 55
I NH+ NH P+ + + RLPP +R + + + L + + P
Sbjct: 387 IQNHNHNHPPEFINHFRHHRLPPPPLSLRSTNLHRLMILQRLQWYLLLQRR*WYLSPLPA 566
Query: 56 DDSTSAEVISDNADNGGERNGGTGKRPAWNRSSGNGGVSEVQPVMDAHSWPALS 109
++ I+ + +GG N G K+PAWN+ NG E+ PVM A SWPALS
Sbjct: 567 PMEKNSNTIASDGGDGG--NAGGSKKPAWNKQPLNGVAVEIGPVMGAESWPALS 722
>TC76311 homologue to GP|3204132|emb|CAA07235.1 extensin {Cicer arietinum},
partial (77%)
Length = 973
Score = 42.0 bits (97), Expect = 6e-04
Identities = 32/101 (31%), Positives = 41/101 (39%), Gaps = 8/101 (7%)
Frame = +2
Query: 282 PPRVSP----RIIRPSLPPNSAPFIH---PPPLRPFGGHMGFHELAAPVVLFAGPPPPID 334
PP SP + P P +S P H PPP P AP ++ PPPP+
Sbjct: 242 PPTASPPPPYHYVSPPPPTSSPPPYHYTSPPPPSP---------APAPTYIYKSPPPPMK 394
Query: 335 SLRGVPFVPPMPLY-YAGPDPQLHSKIVNQIDYYFSNENLV 374
S PP P+Y YA P P ++ N + N LV
Sbjct: 395 S-------PPPPVYIYASPPPPIYK*AQNTTTAFLINHVLV 496
Score = 33.1 bits (74), Expect = 0.27
Identities = 24/75 (32%), Positives = 26/75 (34%)
Frame = +2
Query: 280 HVPPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGV 339
H PP SP PP + PPP P P + PPPP S
Sbjct: 38 HSPPPPSPS------PPPPYYYKSPPPPSPS---------PPPPYYYKSPPPPSPS---- 160
Query: 340 PFVPPMPLYYAGPDP 354
PP P YY P P
Sbjct: 161 ---PPPPYYYQSPPP 196
>TC76430 similar to PIR|T07623|T07623 extensin homolog HRGP2 - soybean
(fragment), partial (61%)
Length = 912
Score = 40.0 bits (92), Expect = 0.002
Identities = 30/97 (30%), Positives = 39/97 (39%), Gaps = 8/97 (8%)
Frame = +3
Query: 282 PPRVSP----RIIRPSLPPNSAPFIH---PPPLRPFGGHMGFHELAAPVVLFAGPPPPID 334
PP SP + P P +S P H PPP P AP ++ PPPP+
Sbjct: 129 PPTASPPPPYHYVSPPPPTSSPPPYHYTSPPPPSP---------APAPTYIYKSPPPPMK 281
Query: 335 SLRGVPFVPPMPLY-YAGPDPQLHSKIVNQIDYYFSN 370
S PP P+Y YA P P ++ N + N
Sbjct: 282 S-------PPPPVYIYASPPPPIYK*AQNTTTAFLIN 371
>AL376214 homologue to GP|3204132|emb| extensin {Cicer arietinum}, partial
(48%)
Length = 362
Score = 39.3 bits (90), Expect = 0.004
Identities = 27/85 (31%), Positives = 36/85 (41%), Gaps = 5/85 (5%)
Frame = +3
Query: 295 PPNSAP----FIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFVPPMPLY-Y 349
PP S+P + PPP P AP ++ PPPP+ S PP P+Y Y
Sbjct: 123 PPTSSPPPYHYTSPPPPSP---------APAPTYIYKSPPPPMKS-------PPPPVYIY 254
Query: 350 AGPDPQLHSKIVNQIDYYFSNENLV 374
A P P ++ N + N LV
Sbjct: 255 ASPPPPIYK*AQNTTTAFLINHVLV 329
Score = 28.1 bits (61), Expect = 8.6
Identities = 18/51 (35%), Positives = 21/51 (40%)
Frame = +3
Query: 283 PRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPI 333
P +P I S PP P PPP PV ++A PPPPI
Sbjct: 177 PAPAPTYIYKSPPP---PMKSPPP---------------PVYIYASPPPPI 275
>TC86671 similar to PIR|T06782|T06782 extensin - soybean, partial (41%)
Length = 991
Score = 38.9 bits (89), Expect = 0.005
Identities = 23/77 (29%), Positives = 33/77 (41%), Gaps = 1/77 (1%)
Frame = +3
Query: 282 PPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF 341
PP PS PP + PPP P + + P+ ++ PPPPI + +P
Sbjct: 171 PPPPKKPYKYPSPPPQIYKYKSPPPPLPKKPYK-YPSPPPPIYMYKSPPPPIYKYKSLPP 347
Query: 342 VPP-MPLYYAGPDPQLH 357
PP P Y P P ++
Sbjct: 348 PPPKKPYKYPSPPPPVY 398
>TC76431 similar to GP|8132441|gb|AAF73291.1| extensin {Pisum sativum},
partial (30%)
Length = 890
Score = 38.9 bits (89), Expect = 0.005
Identities = 24/89 (26%), Positives = 36/89 (39%), Gaps = 11/89 (12%)
Frame = +1
Query: 280 HVPPRVSPRIIRPSLPPN-SAPFIHPPPLRPFGGHMG-----FHELAAPVVLFAGPPPPI 333
H P P + S PP+ P+ +P P P H +H P ++ PPPP+
Sbjct: 61 HPHPHPHPYPVHHSPPPSPKKPYKYPSPPPPTHHHYPHPHPVYHSPPPPKYKYSSPPPPV 240
Query: 334 DSLRGVPFV-----PPMPLYYAGPDPQLH 357
+ P+ P P Y+ P P +H
Sbjct: 241 HHVSPKPYYHSPPPPKKPYKYSSPPPPVH 327
>BQ144302 similar to GP|6815109|dbj| hypothetical protein {Oryza sativa
(japonica cultivar-group)}, partial (3%)
Length = 1047
Score = 37.7 bits (86), Expect = 0.011
Identities = 19/54 (35%), Positives = 26/54 (47%)
Frame = -2
Query: 70 NGGERNGGTGKRPAWNRSSGNGGVSEVQPVMDAHSWPALSDSARGSTKSESSKG 123
NGGE+NGG G W R G G E+ + + + RG TK ++ KG
Sbjct: 524 NGGEKNGGGGNGEIWERRVGGGSNGEIIKLKEKQG-QTVWKKIRGETKRKNGKG 366
Score = 28.5 bits (62), Expect = 6.6
Identities = 25/59 (42%), Positives = 30/59 (50%)
Frame = +1
Query: 288 RIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFVPPMP 346
++ R PPN PF PPP PF + F L V +FA P I+SL P VPP P
Sbjct: 214 KLSRSIYPPNPFPFHFPPPF-PF---LSF--LRRSVFVFA-PFSFINSL-APPLVPPPP 366
>TC87476 homologue to PIR|T11622|T11622 extensin class 1 precursor - cowpea,
partial (48%)
Length = 939
Score = 37.4 bits (85), Expect = 0.014
Identities = 30/108 (27%), Positives = 39/108 (35%), Gaps = 8/108 (7%)
Frame = +3
Query: 255 HHHHNYGNRRDQDWNSRRNYNGRDMHVPPRVSPR---IIRPSLPPNSAP-----FIHPPP 306
HH H + N R + H PP SP + + PP+ +P + PPP
Sbjct: 288 HHLHTFINLLLHHLLHHRLHISTSHHRPPSPSPPPPYVYKSPPPPSPSPPPPYVYKSPPP 467
Query: 307 LRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFVPPMPLYYAGPDP 354
P P ++ PPPP S PP P YY P P
Sbjct: 468 PSPS---------PPPPYVYKSPPPPSAS-------PPPPYYYKSPPP 563
>TC76721 homologue to GP|8132441|gb|AAF73291.1| extensin {Pisum sativum},
partial (46%)
Length = 674
Score = 36.6 bits (83), Expect = 0.024
Identities = 22/78 (28%), Positives = 29/78 (36%)
Frame = +1
Query: 280 HVPPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGV 339
H PP PS PP + PPP + PV + PPPP+ +
Sbjct: 133 HSPPPPKKPYKYPSPPPPVYKYKSPPP-----PVYKYKSPPPPVYKYKSPPPPVYKYKSP 297
Query: 340 PFVPPMPLYYAGPDPQLH 357
P P P Y P P ++
Sbjct: 298 PPPPKKPYKYPSPPPPVY 351
Score = 31.6 bits (70), Expect = 0.78
Identities = 23/78 (29%), Positives = 32/78 (40%), Gaps = 14/78 (17%)
Frame = +1
Query: 295 PPNSAPFIHP---PPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFV---PPMPLY 348
PP P+ +P PP+ + PV + PPPP+ S +V PP P+Y
Sbjct: 301 PPPKKPYKYPSPPPPVYKYKSPPPPVYSPPPVYKYKSPPPPVYSPPPPHYVYSSPPPPVY 480
Query: 349 --------YAGPDPQLHS 358
Y+ P P HS
Sbjct: 481 SPPPPHYIYSSPPPPYHS 534
>BF642399 similar to PIR|T10863|T10 extensin precursor - kidney bean, partial
(33%)
Length = 653
Score = 36.6 bits (83), Expect = 0.024
Identities = 23/78 (29%), Positives = 32/78 (40%)
Frame = +3
Query: 280 HVPPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGV 339
H PP P + P PP PPP +P+ + PV + PPPP+ +
Sbjct: 138 HSPP---PPVHSPP-PPYHYSSPPPPPKKPYK----YASPPPPVYKYKSPPPPVYKYKSP 293
Query: 340 PFVPPMPLYYAGPDPQLH 357
P P P Y P P ++
Sbjct: 294 PPPPKKPYKYPSPPPPVY 347
Score = 34.7 bits (78), Expect = 0.092
Identities = 16/54 (29%), Positives = 24/54 (43%)
Frame = +3
Query: 304 PPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFVPPMPLYYAGPDPQLH 357
PPP +P+ + PV + PPPP+ + P P P Y P P ++
Sbjct: 297 PPPKKPYK----YPSPPPPVYKYKSPPPPVYKYKSPPPPPKKPYKYPSPPPPVY 446
Score = 33.5 bits (75), Expect = 0.21
Identities = 19/64 (29%), Positives = 25/64 (38%)
Frame = +3
Query: 295 PPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFVPPMPLYYAGPDP 354
PP P+ +P P P + PV + PPPP P PP Y P P
Sbjct: 297 PPPKKPYKYPSPPPPV---YKYKSPPPPVYKYKSPPPPPKKPYKYPSPPPPVYKYKSPPP 467
Query: 355 QLHS 358
++S
Sbjct: 468 PVYS 479
Score = 32.7 bits (73), Expect = 0.35
Identities = 19/64 (29%), Positives = 25/64 (38%), Gaps = 4/64 (6%)
Frame = +3
Query: 295 PPNSAPFIHPPPLRPFGGHMG----FHELAAPVVLFAGPPPPIDSLRGVPFVPPMPLYYA 350
PP P+ +P P P + + PV + PPPP+ S PP P Y
Sbjct: 396 PPPKKPYKYPSPPPPVYKYKSPPPPVYSPPPPVYKYKSPPPPVHS-------PPXPYKYK 554
Query: 351 GPDP 354
P P
Sbjct: 555 SPPP 566
Score = 29.6 bits (65), Expect = 3.0
Identities = 21/79 (26%), Positives = 28/79 (34%)
Frame = +3
Query: 279 MHVPPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRG 338
+H PP P PP P+ + P P + PV + PPPP
Sbjct: 156 VHSPP--PPYHYSSPPPPPKKPYKYASPPPPV---YKYKSPPPPVYKYKSPPPPPKKPYK 320
Query: 339 VPFVPPMPLYYAGPDPQLH 357
P PP Y P P ++
Sbjct: 321 YPSPPPPVYKYKSPPPPVY 377
Score = 29.3 bits (64), Expect = 3.9
Identities = 16/51 (31%), Positives = 21/51 (40%)
Frame = +3
Query: 304 PPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFVPPMPLYYAGPDP 354
PPP +P+ H PPPP+ S PP P +Y+ P P
Sbjct: 111 PPPPKPYYYH--------------SPPPPVHS-------PPPPYHYSSPPP 200
Score = 28.5 bits (62), Expect = 6.6
Identities = 13/41 (31%), Positives = 19/41 (45%)
Frame = +3
Query: 318 ELAAPVVLFAGPPPPIDSLRGVPFVPPMPLYYAGPDPQLHS 358
+ +A +++ PPPP P P YY P P +HS
Sbjct: 75 QTSANSYIYSSPPPP-----------PKPYYYHSPPPPVHS 164
>TC87240 similar to GP|19571126|dbj|BAB86550. putative 5-3 exoribonuclease
{Oryza sativa (japonica cultivar-group)}, partial (43%)
Length = 2400
Score = 36.6 bits (83), Expect = 0.024
Identities = 26/100 (26%), Positives = 40/100 (40%)
Frame = +1
Query: 177 GHTQQSAPQVSIAATGSHTSSSRDHTQSPRDHTQSPRDHAQSPRDHTQRSGFVPSDHPQQ 236
G+ Q +P V + H++S Q+PR H H + + SG + +P+
Sbjct: 1756 GYNQPYSPPVIPSPHQHHSNSFPRGDQNPRSHHSDRNSHHVNGGNARHSSG--NNQNPRF 1929
Query: 237 RNSFRHRNGGPHQRGDGSHHHHNYGNRRDQDWNSRRNYNG 276
N+ H G H+ H R DQ W R N +G
Sbjct: 1930 SNTQGSFPRQGHHHDAGRHNQHFQPYRADQHWTPRGNPSG 2049
>TC76718 homologue to GP|8132441|gb|AAF73291.1| extensin {Pisum sativum},
partial (63%)
Length = 712
Score = 36.6 bits (83), Expect = 0.024
Identities = 23/78 (29%), Positives = 32/78 (40%)
Frame = +1
Query: 280 HVPPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGV 339
H PP P + P PP PPP +P+ + PV + PPPP+ +
Sbjct: 163 HSPP---PPVHSPP-PPYHYSSPPPPPKKPYK----YASPPPPVYKYKSPPPPVYKYKSP 318
Query: 340 PFVPPMPLYYAGPDPQLH 357
P P P Y P P ++
Sbjct: 319 PPPPKKPYKYPSPPPPVY 372
Score = 33.5 bits (75), Expect = 0.21
Identities = 22/81 (27%), Positives = 29/81 (35%), Gaps = 5/81 (6%)
Frame = +1
Query: 282 PPRVSPRIIRPSLPPNSAPFIHPPP-----LRPFGGHMGFHELAAPVVLFAGPPPPIDSL 336
PP PS PP + PPP P + PV + PPPP+
Sbjct: 319 PPPPKKPYKYPSPPPPVYKYKSPPPPVYKYKSPPPPVYKYKSPPPPVYKYKSPPPPVYKY 498
Query: 337 RGVPFVPPMPLYYAGPDPQLH 357
+ P P P Y P P ++
Sbjct: 499 KSPPPPPKKPYKYPSPPPPVY 561
Score = 29.6 bits (65), Expect = 3.0
Identities = 21/79 (26%), Positives = 28/79 (34%)
Frame = +1
Query: 279 MHVPPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRG 338
+H PP P PP P+ + P P + PV + PPPP
Sbjct: 181 VHSPP--PPYHYSSPPPPPKKPYKYASPPPPV---YKYKSPPPPVYKYKSPPPPPKKPYK 345
Query: 339 VPFVPPMPLYYAGPDPQLH 357
P PP Y P P ++
Sbjct: 346 YPSPPPPVYKYKSPPPPVY 402
Score = 29.3 bits (64), Expect = 3.9
Identities = 16/51 (31%), Positives = 21/51 (40%)
Frame = +1
Query: 304 PPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFVPPMPLYYAGPDP 354
PPP +P+ H PPPP+ S PP P +Y+ P P
Sbjct: 136 PPPPKPYYYH--------------SPPPPVHS-------PPPPYHYSSPPP 225
Score = 28.5 bits (62), Expect = 6.6
Identities = 13/41 (31%), Positives = 19/41 (45%)
Frame = +1
Query: 318 ELAAPVVLFAGPPPPIDSLRGVPFVPPMPLYYAGPDPQLHS 358
+ +A +++ PPPP P P YY P P +HS
Sbjct: 100 QTSANSYIYSSPPPP-----------PKPYYYHSPPPPVHS 189
>TC76350 similar to GP|8132441|gb|AAF73291.1| extensin {Pisum sativum},
partial (63%)
Length = 1008
Score = 36.2 bits (82), Expect = 0.032
Identities = 19/56 (33%), Positives = 28/56 (49%), Gaps = 1/56 (1%)
Frame = +2
Query: 304 PPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFVPPMPLY-YAGPDPQLHS 358
PPP +P+ + PV + PPPP+ S + PP P+Y Y P P ++S
Sbjct: 146 PPPSKPYK----YASPPPPVYKYKSPPPPVYSPPPPVYSPPPPVYKYKSPPPPVYS 301
Score = 31.2 bits (69), Expect = 1.0
Identities = 23/78 (29%), Positives = 31/78 (39%), Gaps = 1/78 (1%)
Frame = +2
Query: 282 PPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF 341
PP SP PP + PPP + PV + PPPP+ +
Sbjct: 287 PPVYSP-------PPPVYKYKSPPP--------PVYSPPPPVYKYKSPPPPV-------Y 400
Query: 342 VPPMPLY-YAGPDPQLHS 358
PP P+Y Y P P ++S
Sbjct: 401 SPPPPVYKYKSPPPPVYS 454
Score = 31.2 bits (69), Expect = 1.0
Identities = 23/78 (29%), Positives = 31/78 (39%), Gaps = 1/78 (1%)
Frame = +2
Query: 282 PPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF 341
PP SP PP + PPP + PV + PPPP+ +
Sbjct: 389 PPVYSP-------PPPVYKYKSPPP--------PVYSPPPPVYKYKSPPPPV-------Y 502
Query: 342 VPPMPLY-YAGPDPQLHS 358
PP P+Y Y P P ++S
Sbjct: 503 SPPPPVYKYKSPPPPVYS 556
Score = 30.4 bits (67), Expect = 1.7
Identities = 23/78 (29%), Positives = 30/78 (37%), Gaps = 1/78 (1%)
Frame = +2
Query: 282 PPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF 341
PP SP PP + PPP + PV + PPPP+ +
Sbjct: 491 PPVYSP-------PPPVYKYKSPPP--------PVYSPPPPVYKYKSPPPPV-------Y 604
Query: 342 VPPMPLY-YAGPDPQLHS 358
PP P+Y Y P P +S
Sbjct: 605 SPPPPVYKYKSPPPPAYS 658
>TC87201 similar to PIR|F86388|F86388 hypothetical protein AAG29221.1
[imported] - Arabidopsis thaliana, partial (41%)
Length = 991
Score = 35.8 bits (81), Expect = 0.041
Identities = 23/75 (30%), Positives = 31/75 (40%), Gaps = 2/75 (2%)
Frame = +1
Query: 282 PPRVSPRIIRPSLPPNSAPFIHP--PPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGV 339
PP P IR + + P+++P PP P ++ P V + PPPP
Sbjct: 55 PPPSPPPPIRIQITTRAPPYVYPSPPPPSPPPPYIYKSPPPPPYVYKSPPPPPYVYPSPP 234
Query: 340 PFVPPMPLYYAGPDP 354
P PP P Y P P
Sbjct: 235 PPSPPPPYIYKSPPP 279
Score = 32.0 bits (71), Expect = 0.60
Identities = 21/69 (30%), Positives = 28/69 (40%)
Frame = +1
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFVPPM 345
S ++RP + P+ P PPP+R AP ++ PPPP PP
Sbjct: 16 SHHLLRPYVYPSPPPPSPPPPIRI------QITTRAPPYVYPSPPPP---------SPPP 150
Query: 346 PLYYAGPDP 354
P Y P P
Sbjct: 151 PYIYKSPPP 177
>BQ144079 similar to GP|6523547|emb| hydroxyproline-rich glycoprotein DZ-HRGP
{Volvox carteri f. nagariensis}, partial (12%)
Length = 1071
Score = 35.8 bits (81), Expect = 0.041
Identities = 37/176 (21%), Positives = 63/176 (35%), Gaps = 13/176 (7%)
Frame = +3
Query: 198 SRDHTQSPRDHTQSPRDHAQSPRD-----HTQRSGFVPSDHPQQRNSFRHRNGGPHQRGD 252
S ++T P + + R H + H+ + H Q + + GP +
Sbjct: 321 SANNTTKPTNEPTNLRTHFRQTHPPIYDIHSPPGTYHSQAHTQPQPFASQKPNGPCKHSA 500
Query: 253 GSHHHHNYGNRRDQDWNSRRNYNGRDMHVPP--RVSPRIIRPSLPPN-----SAPFIHPP 305
H H + D + + N P + +P I P+ PPN SA I P
Sbjct: 501 TEHGQHKHKQAADPP-HPKPPTNPPKPPTPQCAKTNPNPIPPATPPNPPPPRSATKIRHP 677
Query: 306 PLRPFGGHMGFHELA-APVVLFAGPPPPIDSLRGVPFVPPMPLYYAGPDPQLHSKI 360
+P H + +P + PPP +P PP P P Q+++++
Sbjct: 678 TAKPLPQETQSHPIPESPNTPHSHTPPPRPPNPPIPHYPPRPYRANAPPRQIYARM 845
>TC79235 weakly similar to GP|7297741|gb|AAF52992.1| CG17108 gene product
{Drosophila melanogaster}, partial (21%)
Length = 648
Score = 35.8 bits (81), Expect = 0.041
Identities = 25/82 (30%), Positives = 30/82 (36%)
Frame = -2
Query: 273 NYNGRDMHVPPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPP 332
+Y H PP P I+ P P PPP P H PPPP
Sbjct: 536 SYPHNHRHNPPPPGPPIMGRGPPLGPGP---PPPPMPHSHH---------------PPPP 411
Query: 333 IDSLRGVPFVPPMPLYYAGPDP 354
D PF PP P+++ P P
Sbjct: 410 HD-----PFAPPPPVFHHPPPP 360
Score = 35.4 bits (80), Expect = 0.054
Identities = 25/76 (32%), Positives = 28/76 (35%), Gaps = 8/76 (10%)
Frame = -2
Query: 282 PPRVSPRIIRPSLPPNSAPFIHPPPL--------RPFGGHMGFHELAAPVVLFAGPPPPI 333
PP P P PP PF PPP+ P+ H P+ PPPP
Sbjct: 452 PPPPMPHSHHP--PPPHDPFAPPPPVFHHPPPPPNPYAPPPVVHHHHTPLHAHPAPPPPQ 279
Query: 334 DSLRGVPFVPPMPLYY 349
G P PP P YY
Sbjct: 278 PGHPGPP--PPRPPYY 237
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.312 0.130 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,046,844
Number of Sequences: 36976
Number of extensions: 304093
Number of successful extensions: 2518
Number of sequences better than 10.0: 187
Number of HSP's better than 10.0 without gapping: 2080
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2355
length of query: 522
length of database: 9,014,727
effective HSP length: 100
effective length of query: 422
effective length of database: 5,317,127
effective search space: 2243827594
effective search space used: 2243827594
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC123571.8


