

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
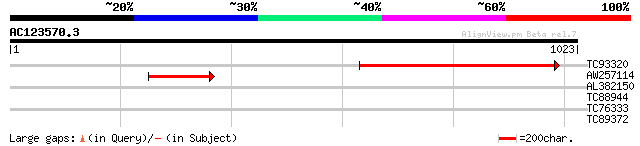
Query= AC123570.3 + phase: 0 /pseudo
(1023 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC93320 490 e-138
AW257114 194 1e-49
AL382150 similar to GP|22534308|gb agglutinin receptor {Streptoc... 33 0.56
TC88944 type IIB calcium ATPase [Medicago truncatula] 32 1.2
TC76333 similar to SP|P49044|VPE_VICSA Vacuolar processing enzym... 32 1.2
TC89372 similar to GP|22324595|gb|AAM95623.1 anaphase promoting ... 29 8.1
>TC93320
Length = 1094
Score = 490 bits (1261), Expect = e-138
Identities = 269/364 (73%), Positives = 299/364 (81%), Gaps = 3/364 (0%)
Frame = +3
Query: 631 LTFEKYDISLRDEGILLLFASVIIALRPKTHVPNIRGILHLFIITLLKGVVPVAQALGSM 690
LT KYDIS RDEGILLLFAS IIALRPKTH+PNIRG+LHLFIITLLKGVVPVAQALGSM
Sbjct: 3 LTSGKYDISPRDEGILLLFASXIIALRPKTHIPNIRGLLHLFIITLLKGVVPVAQALGSM 182
Query: 691 VNKLISKSNGAEKSDELTLEEALHIIFNTKICFSSDNMLQICDGSINRNEIVLTDVCLGM 750
+NKL SKSNGA+KSDELTLEEAL IIFNTKI FSS+NMLQI +GS N ++IVLTD+CLG+
Sbjct: 183 LNKLTSKSNGAKKSDELTLEEALDIIFNTKIWFSSNNMLQIYNGSSNGSDIVLTDLCLGI 362
Query: 751 TNDRLLQTNAVCGLSWIGKGLLLRGHEKIKDITKILTECLISD-RNSSLPLIEGLDENNE 809
TNDRLLQ+NA+CGLSWIGKGLLLRGHEKIKDIT I TECLISD R +S+PL+EG EN E
Sbjct: 363 TNDRLLQSNAICGLSWIGKGLLLRGHEKIKDITMIFTECLISDRRKTSVPLVEGSLENTE 542
Query: 810 EHKGDHLARKCAADAFHVLMSDAEDCLNRKFHATMRPLYKQRFFSSMMPIFLQLISRSDS 869
+ K D LARKCA +AFHVLMSDAEDCLNRKFHAT+RPLYKQRFFSSMMPIFLQLISRSDS
Sbjct: 543 KQKCDPLARKCATEAFHVLMSDAEDCLNRKFHATVRPLYKQRFFSSMMPIFLQLISRSDS 722
Query: 870 SSSRYLLLRAFARVMSVTPLIVILNDAKELISVLLDCLSMLTEDIQDKDILYG-LLLVLS 928
SR LLLRAFA VMS TPLIVILN+AK+LI VLLDCL MLTEDIQDKDILYG + +
Sbjct: 723 LLSRSLLLRAFAHVMSDTPLIVILNEAKKLIPVLLDCLFMLTEDIQDKDILYGSVASFIW 902
Query: 929 GMLTEKNESLLFKAIQIKNDVNG*FLFLPIL-QLVRESCIQCLVALSKLPHVRIYPLRTQ 987
G K + + I + + L ++ +L LVALS+LPHVRIYPLRTQ
Sbjct: 903 GC*QRKMDKKQSSRMHILSSMV*LSLSTTLIRRLFGRQLFNGLVALSELPHVRIYPLRTQ 1082
Query: 988 VLEA 991
V A
Sbjct: 1083VSAA 1094
>AW257114
Length = 360
Score = 194 bits (493), Expect = 1e-49
Identities = 100/120 (83%), Positives = 105/120 (87%)
Frame = +1
Query: 250 FIINSIASYEMYDAVSVQEKKKLHVIGRILYIFAKTSIPSCNAVFQSLLLRMMDSLGFSV 309
FI NSIASYEMYD +SVQEKKKLH IGRILYI AKTSIPSCNAVFQSL LRMMD LGFS
Sbjct: 1 FITNSIASYEMYDTISVQEKKKLHAIGRILYISAKTSIPSCNAVFQSLFLRMMDKLGFSA 180
Query: 310 SNIDGLKNAGILASQSVNFGFLYLCIELLAGCRELVILSEEKPGTCFTILHSSSDFLFNS 369
SNIDGL+N GILASQSVNFGFLYLCIELL+ CRELVILS+EK T TILHSSS LFN+
Sbjct: 181 SNIDGLQNGGILASQSVNFGFLYLCIELLSXCRELVILSDEKRETYCTILHSSSAVLFNA 360
>AL382150 similar to GP|22534308|gb agglutinin receptor {Streptococcus
agalactiae 2603V/R}, partial (0%)
Length = 439
Score = 33.1 bits (74), Expect = 0.56
Identities = 12/33 (36%), Positives = 24/33 (72%)
Frame = -2
Query: 279 LYIFAKTSIPSCNAVFQSLLLRMMDSLGFSVSN 311
+Y F +SIP+CN++ S +L ++DS F++++
Sbjct: 165 MYCFLSSSIPNCNSLLISSILALLDSTSFNLAS 67
>TC88944 type IIB calcium ATPase [Medicago truncatula]
Length = 1883
Score = 32.0 bits (71), Expect = 1.2
Identities = 12/52 (23%), Positives = 32/52 (61%)
Frame = +3
Query: 251 IINSIASYEMYDAVSVQEKKKLHVIGRILYIFAKTSIPSCNAVFQSLLLRMM 302
I NS ++++ +S ++ ++++V IL + T++ +C A+FQ +++ +
Sbjct: 1548 IFNSFVFCQVFNEISSRDMERINVFEGILKNYVFTAVLTCTAIFQIIIVEFL 1703
>TC76333 similar to SP|P49044|VPE_VICSA Vacuolar processing enzyme precursor
(EC 3.4.22.-) (VPE) (Proteinase B). [Spring vetch Tare],
partial (91%)
Length = 2253
Score = 32.0 bits (71), Expect = 1.2
Identities = 18/50 (36%), Positives = 27/50 (54%), Gaps = 2/50 (4%)
Frame = -3
Query: 64 ERDPECLMPAFHIVESLARLYP--DPSGLFASFARDVFDLLEPYFPIQFT 111
+RDP+C+ F +V ++L DP+GL S + F + FPI FT
Sbjct: 1327 DRDPQCVSKVFTMVLRQSQLSTSGDPAGLTLSSSSVPFSMPNKSFPISFT 1178
>TC89372 similar to GP|22324595|gb|AAM95623.1 anaphase promoting
complex/cyclosome subunit {Arabidopsis thaliana},
partial (42%)
Length = 953
Score = 29.3 bits (64), Expect = 8.1
Identities = 14/30 (46%), Positives = 19/30 (62%), Gaps = 3/30 (10%)
Frame = +2
Query: 30 LCFELLDCLLEHH---ADSVASLEEDLIFG 56
LC+E L+CL+E+H D A+L L FG
Sbjct: 614 LCYEALECLIENHMLTCDEEANLISSLQFG 703
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.326 0.141 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,082,831
Number of Sequences: 36976
Number of extensions: 502945
Number of successful extensions: 3012
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 2198
Number of HSP's successfully gapped in prelim test: 138
Number of HSP's that attempted gapping in prelim test: 775
Number of HSP's gapped (non-prelim): 2395
length of query: 1023
length of database: 9,014,727
effective HSP length: 106
effective length of query: 917
effective length of database: 5,095,271
effective search space: 4672363507
effective search space used: 4672363507
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC123570.3


