

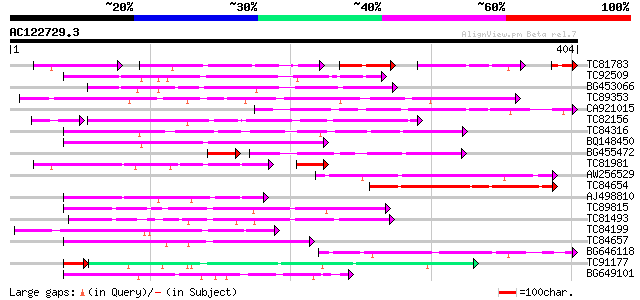
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122729.3 + phase: 0
(404 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC81783 similar to GP|9294656|dbj|BAB03005.1 gb|AAF30317.1~gene_... 77 6e-42
TC92509 weakly similar to PIR|T49129|T49129 hypothetical protein... 144 5e-35
BG453066 138 3e-33
TC89353 weakly similar to GP|9279704|dbj|BAB01261.1 gb|AAF25964.... 130 1e-30
CA921015 homologue to SP|Q8Y7G1|DPO3 DNA polymerase III polC-typ... 129 2e-30
TC82156 similar to GP|20197985|gb|AAM15340.1 F-box protein famil... 114 4e-29
TC84316 weakly similar to GP|11994612|dbj|BAB02749. gb|AAC24186.... 119 2e-27
BQ148450 similar to GP|10177464|dbj gb|AAD21700.1~gene_id:MQB2.1... 117 1e-26
BG455472 101 1e-25
TC81981 similar to GP|11994545|dbj|BAB02732. emb|CAB62106.1~gene... 95 1e-23
AW256529 homologue to GP|496669|emb|C C-169 protein {Saccharomyc... 104 5e-23
TC84654 101 6e-22
AJ498810 weakly similar to PIR|D86339|D863 protein F2D10.14 [imp... 97 1e-20
TC89815 96 2e-20
TC81493 weakly similar to PIR|F86249|F86249 protein F25C20.23 [i... 92 5e-19
TC84199 similar to GP|9294656|dbj|BAB03005.1 gb|AAF30317.1~gene_... 86 3e-17
TC84657 weakly similar to GP|9294656|dbj|BAB03005.1 gb|AAF30317.... 86 3e-17
BG646118 78 5e-15
TC91177 similar to GP|9294656|dbj|BAB03005.1 gb|AAF30317.1~gene_... 72 1e-14
BG649101 homologue to GP|6648199|gb|A unknown protein {Arabidops... 76 2e-14
>TC81783 similar to GP|9294656|dbj|BAB03005.1
gb|AAF30317.1~gene_id:F14O13.6~similar to unknown
protein {Arabidopsis thaliana}, partial (6%)
Length = 1225
Score = 76.6 bits (187), Expect(5) = 6e-42
Identities = 55/140 (39%), Positives = 70/140 (49%), Gaps = 8/140 (5%)
Frame = +3
Query: 93 ELLLVDSRLPSVTAIIPDTTHN--------FRLNPSDNHPIMIDSCDGIICFENRNDNHV 144
+ +L DS +PSV + T R N + I + SCDGI C E N
Sbjct: 315 DFVLHDSPIPSVFSTSTSVTQTQLDPPFTLARRNYVNATAIAMCSCDGIFCGEL---NLG 485
Query: 145 DLVVWNPCTGKFKILPPLENIPNGKTHTLYSIGYDRFVDNYKVVAFSCHRQINKSYKYCN 204
VWNP KFK+LPP +N G ++ S GYD F+DNYKVVA S KY
Sbjct: 486 YCFVWNPSIRKFKLLPPYKNPLEGDPFSI-SFGYDHFIDNYKVVAISS--------KY-- 632
Query: 205 SQVRVHTLGTNFWRRIPNFP 224
+V V+ LGT++WRRI P
Sbjct: 633 -EVFVNALGTDYWRRIREHP 689
Score = 60.8 bits (146), Expect(5) = 6e-42
Identities = 32/83 (38%), Positives = 49/83 (58%), Gaps = 6/83 (7%)
Frame = +2
Query: 291 KDCLCVLVYTETLLGIWVMKDYGNKNSWTKLFAVPYAKVGYHGFGFVDLHYISEEDDQVF 350
+DCLCV ++ +W+MK+YGN+ SWTKL++VP K+ + F + YI EDDQ+
Sbjct: 881 RDCLCVYEASDLFFNVWIMKEYGNQESWTKLYSVP--KMQHRRFKAYTISYI-YEDDQLL 1051
Query: 351 LHFC------SKVYVYNYKNSTV 367
L +K+ VY+ K T+
Sbjct: 1052LGIVDMQSSNTKLAVYDSKTGTL 1120
Score = 51.6 bits (122), Expect(5) = 6e-42
Identities = 31/68 (45%), Positives = 37/68 (53%), Gaps = 5/68 (7%)
Frame = +2
Query: 18 STTVRKQNSSD-----PFGLFALLWLDLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKF 72
+T R+Q S+ P L DL EIL RLPVK L+ L+C+ K NSLISDP F
Sbjct: 65 ATRTRRQRSNGTLTYPPEPELPTLPFDLLPEILCRLPVKLLIQLRCLCKFFNSLISDPNF 244
Query: 73 VKHHLHLS 80
K H S
Sbjct: 245 AKKHFQFS 268
Score = 37.0 bits (84), Expect(5) = 6e-42
Identities = 20/40 (50%), Positives = 24/40 (60%)
Frame = +1
Query: 236 GKFVSGTINWAIENQKNYDSWVILSLDLGNESYQEISRPD 275
G FV GT+NW + D ILSLDL ESYQ++ PD
Sbjct: 715 GIFVGGTVNWMASDIA--DLLFILSLDLEKESYQKLFLPD 828
Score = 23.5 bits (49), Expect(5) = 6e-42
Identities = 11/18 (61%), Positives = 14/18 (77%)
Frame = +1
Query: 387 FESLYDSSGVYVESLISP 404
+E +Y S VY+ESLISP
Sbjct: 1144 YEPIY--SNVYIESLISP 1191
>TC92509 weakly similar to PIR|T49129|T49129 hypothetical protein F26G5.80 -
Arabidopsis thaliana, partial (8%)
Length = 858
Score = 144 bits (364), Expect = 5e-35
Identities = 101/243 (41%), Positives = 136/243 (55%), Gaps = 13/243 (5%)
Frame = +3
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLIRNS----EL 94
+L EIL RLPVK L L+C+ KS NSLISDPKF K HLH S T P+HL++R++
Sbjct: 183 ELVAEILCRLPVKLLXQLRCLCKSFNSLISDPKFAKKHLH-SSTTPHHLILRSNNGSGRF 359
Query: 95 LLVDSRLPSV----TAIIPDT--THNFRLNPSDNHPIMIDSCDGIICFENRNDNHVDLVV 148
L+ S + SV T +P T T+ L P SCDGIIC ++ V+
Sbjct: 360 ALIVSPIQSVLSTSTVPVPQTQLTYPTCLTEEFASPYEWCSCDGIICL---TTDYSSAVL 530
Query: 149 WNPCTGKFKILPPLENIPNGKTHT-LYSIGYDRFVDNYKVVAFSCHRQINKSYKYC--NS 205
WNP KFK LPPL+ I ++ + L++ GYD F DNYKV A + +C +
Sbjct: 531 WNPFINKFKTLPPLKYISLKRSPSCLFTFGYDPFADNYKVFAIT----------FCVKRT 680
Query: 206 QVRVHTLGTNFWRRIPNFPSNIMGLPNGYVGKFVSGTINWAIENQKNYDSWVILSLDLGN 265
V VHT+GT+ WRRI +FPS +P+ G FV+G ++W + I+SLDL +
Sbjct: 681 TVEVHTMGTSSWRRIEDFPSWSF-IPDS--GIFVAGYVHWLTYDGPG-SQREIVSLDLED 848
Query: 266 ESY 268
ESY
Sbjct: 849 ESY 857
>BG453066
Length = 655
Score = 138 bits (348), Expect = 3e-33
Identities = 89/232 (38%), Positives = 125/232 (53%), Gaps = 11/232 (4%)
Frame = +1
Query: 56 LKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLI----RNSELLLVDSRLPSV--TAIIP 109
L+C+ K NSLISDPKFVK HL S T+ HL++ + ++ DS +PS+ T+ I
Sbjct: 1 LRCLCKFFNSLISDPKFVKKHLQ-SSTKRRHLMLTTIDHQQQFVMYDSPIPSLFSTSTIV 177
Query: 110 DTTHNFRLNPSDNHPIMIDSCDGIICFENRNDNHVDLVVWNPCTGKFKILPPLENIPNGK 169
T + N D + + SCDGI +H +WNP KFK+LPPLEN
Sbjct: 178 AQTQLYPPN-GDTYASVKCSCDGIFL---GMFSHTSYFLWNPSIRKFKLLPPLENQDKSA 345
Query: 170 THTLY-----SIGYDRFVDNYKVVAFSCHRQINKSYKYCNSQVRVHTLGTNFWRRIPNFP 224
T+ S GYD F+D YKV+A S ++V ++TLGT++W+RI + P
Sbjct: 346 PFTIIVPYTISFGYDCFIDKYKVIAVSS-----------KNEVFLYTLGTDYWKRIDDIP 492
Query: 225 SNIMGLPNGYVGKFVSGTINWAIENQKNYDSWVILSLDLGNESYQEISRPDF 276
+G +VSG +NW + ++ + ILSLDL NESYQE+ PDF
Sbjct: 493 YYCTICRSGL---YVSGAVNWYVHDESXSSLYFILSLDLXNESYQELYPPDF 639
>TC89353 weakly similar to GP|9279704|dbj|BAB01261.1
gb|AAF25964.1~gene_id:MYA6.2~similar to unknown protein
{Arabidopsis thaliana}, partial (10%)
Length = 1482
Score = 130 bits (326), Expect = 1e-30
Identities = 115/375 (30%), Positives = 176/375 (46%), Gaps = 18/375 (4%)
Frame = +3
Query: 8 SKRKHRHNVVSTTVRKQNSSDPFGLFALLWLDLFLEILYRLPVKSLLVLKCVSKSLNSLI 67
++R+ R + + TV Q+ S+ + A + ++ +EIL RLPV+SLL +CV K +LI
Sbjct: 30 TRRRLRQHSQNVTVFTQSVSE---ITADMPEEIIVEILLRLPVRSLLQFRCVCKLWKTLI 200
Query: 68 SDPKFVKHHLHLSQTRP--YHLLIRNSELLLVDSRLPSVTAIIPDTTHNFRLNPSDNHPI 125
SDP+F K H+ +S P + + ++ LV L + D R+ P+D I
Sbjct: 201 SDPQFAKKHVSISTAYPQLVSVFVSIAKCNLVSYPLKPLL----DNPSAHRVEPADFEMI 368
Query: 126 ------MIDSCDGIICFENRNDNHVDLVVWNPCTGKFKILPPLENIP----NGKTHTLYS 175
+I SC+G++C + +WNP K K P I + K
Sbjct: 369 HTTSMTIIGSCNGLLCL----SDFYQFTLWNPSI-KLKSKPSPTIIAFDSFDSKRFLYRG 533
Query: 176 IGYDRFVDNYKVVAFSCHRQINKSYKYCNSQVRVHTLGTNFWRRIPNFPSNIMGLPNGY- 234
GYD+ D YKV+A + Y ++ ++T G W I FP + G
Sbjct: 534 FGYDQVNDRYKVLAV-----VQNCYNLDETKTLIYTFGGKDWTTIQKFPCDPSRCDLGRL 698
Query: 235 -VGKFVSGTINWAIENQKNYDSWVILSLDLGNESYQEISRP-DFGLDDPVHIFTLGVSKD 292
VGKFVSG +NW + + VI+ D+ E+Y E+S P D+G + V L VS +
Sbjct: 699 GVGKFVSGNLNWIVSKK------VIVFFDIEKETYGEMSLPQDYGDKNTV----LYVSSN 848
Query: 293 CLCVLV--YTETLLGIWVMKDYGNKNSWTKLFAVPYAKVGYHG-FGFVDLHYISEEDDQV 349
+ V +T +W+MK+YG SWTKL +P K+ G + D +ISE +
Sbjct: 849 RIYVSFDHSNKTHWVVWMMKEYGVVESWTKLMIIPQDKLTSPGPYCLSDALFISEHGVLL 1028
Query: 350 FLHFCSKVYVYNYKN 364
SK+ VYN N
Sbjct: 1029MRPQHSKLSVYNLNN 1073
>CA921015 homologue to SP|Q8Y7G1|DPO3 DNA polymerase III polC-type (EC
2.7.7.7) (PolIII). {Listeria monocytogenes}, partial
(0%)
Length = 824
Score = 129 bits (325), Expect = 2e-30
Identities = 85/238 (35%), Positives = 123/238 (50%), Gaps = 8/238 (3%)
Frame = -1
Query: 175 SIGYDRFVDNYKVVAFSCHRQINKSYKYCNSQVRVHTLGTNFWRRIPNFPSNIMGLPNGY 234
+ GYD F+DN +V+ + ++V V+TLGT++W+RI + P NI
Sbjct: 824 NFGYDHFIDNIRVM-----------FVLPKNEVSVYTLGTDYWKRIEDIPYNIFE----- 693
Query: 235 VGKFVSGTINWAIENQKNYDSWVILSLDLGNESYQEISRPDFGLDDPVHIFTLGVSKDCL 294
G FVSGT+NW D ILSLD+ ESYQ++ PD D ++ LGV ++CL
Sbjct: 692 EGVFVSGTVNWLAS-----DDSFILSLDVEKESYQQVLLPDTEND----LWILGVLRNCL 540
Query: 295 CVLVYTETLLGIWVMKDYGNKNSWTKLFAVPYAKVGYHGFGFVDLHYISEEDDQVFLHFC 354
C+L + L +W+M +YGN+ SWTKL++VP + HG + Y S EDDQ+ L F
Sbjct: 539 CILATSNLFLDVWIMNEYGNQESWTKLYSVP--NMQDHGLEAYTVLY-SSEDDQLLLEFN 369
Query: 355 S------KVYVYNYKNSTVKTLDIQGQPSILYNSSRVYFESLYDS--SGVYVESLISP 404
K+ VY+ K T+ + F++ YD Y+ESLISP
Sbjct: 368 EMRSDKVKLVVYDSKTGTLNIPE---------------FQNNYDQICQNFYIESLISP 240
>TC82156 similar to GP|20197985|gb|AAM15340.1 F-box protein family AtFBX9
{Arabidopsis thaliana}, partial (4%)
Length = 1136
Score = 114 bits (286), Expect(2) = 4e-29
Identities = 90/243 (37%), Positives = 126/243 (51%), Gaps = 4/243 (1%)
Frame = +3
Query: 56 LKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLIRNSELLLVDSRLPSVTAIIPDTTHNF 115
+K VSKS SLISD F K +L +S T L + ++ + + + I T
Sbjct: 456 VKSVSKSWKSLISDSNFTKKNLRVSTTSHRLLFPKLTKGQYIFNACTLSSLITTKGTATA 635
Query: 116 RLNPSDNHPI--MIDSCDGIICFENRNDNHVDLVVWNPCTGKFKILPPLENIPNGKTHTL 173
+P + + SC GI+C E ++WNP K+ LPPLE IP ++T+
Sbjct: 636 MQHPLNIRKFDKIRGSCHGILCLELHQRF---AILWNPFINKYASLPPLE-IP--WSNTI 797
Query: 174 YS-IGYDRFVDNYKVVAFSCHRQINKSYKYCNSQVRVHTLGTNFWRRIPNFPSNIMGLPN 232
YS GYD D+YKV AF ++ YK VHT+GT WR I +FP P
Sbjct: 798 YSCFGYDHSTDSYKVAAFIKWMPNSEIYK-----TYVHTMGTTSWRMIQDFPCT----PY 950
Query: 233 GYVGKFVSGTINW-AIENQKNYDSWVILSLDLGNESYQEISRPDFGLDDPVHIFTLGVSK 291
GKFVS T NW A +++ + S +++SL L NESY EI +PD+G D + I +L V +
Sbjct: 951 LKSGKFVSWTFNWLAYKDKYSVSSLLVVSLHLENESYGEILQPDYGSVDVLSI-SLWVLR 1127
Query: 292 DCL 294
DCL
Sbjct: 1128DCL 1136
Score = 31.2 bits (69), Expect(2) = 4e-29
Identities = 20/38 (52%), Positives = 22/38 (57%)
Frame = +2
Query: 16 VVSTTVRKQNSSDPFGLFALLWLDLFLEILYRLPVKSL 53
V TTV K S+P L DL +ILYRLPVKSL
Sbjct: 350 VSRTTVTKSRRSNP-----TLPFDLVEDILYRLPVKSL 448
>TC84316 weakly similar to GP|11994612|dbj|BAB02749.
gb|AAC24186.1~gene_id:MGL6.4~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (10%)
Length = 892
Score = 119 bits (298), Expect = 2e-27
Identities = 90/304 (29%), Positives = 143/304 (46%), Gaps = 16/304 (5%)
Frame = +3
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLIRN------- 91
+L + IL RLPV+SLL KCV KS +L SD F +H +S P + +
Sbjct: 51 ELIVIILLRLPVRSLLRFKCVCKSWKTLFSDTHFANNHFLISTVYPQLVACESVSAYRTW 230
Query: 92 -SELLLVDSRLP-SVTAIIPDTTHNFRLNPSDNHPIMIDSCDGIICFENRNDNHVDLVVW 149
+ ++S L S T +IP + N ++ SC+G +C + V L W
Sbjct: 231 EIKTYPIESLLENSSTTVIPVS------NTGHQRYTILGSCNGFLCLYDNYQRCVRL--W 386
Query: 150 NPCTG-KFKILPPLENIPNGKTHTLYSIGYDRFVDNYKVVAFSCHRQINKSYKYCNSQVR 208
NP K K P ++ Y GYD+ YK++A +I ++
Sbjct: 387 NPSINLKSKSSPTIDRF------IYYGFGYDQVNHKYKLLAVKAFSRI--------TETM 524
Query: 209 VHTLGTNFWRRI-----PNFPSNIMGLPNGYVGKFVSGTINWAIENQKNYDSWVILSLDL 263
++T G N + + P +P N ++GKFVSGT+NW ++ + + ILS D+
Sbjct: 525 IYTFGENSCKNVEVKDFPRYPPN-----RKHLGKFVSGTLNWIVDERDGRAT--ILSFDI 683
Query: 264 GNESYQEISRPDFGLDDPVHIFTLGVSKDCLCVLV-YTETLLGIWVMKDYGNKNSWTKLF 322
E+Y+++ P G V+ L V +C+CV + +T +W+MK YG SWTKL
Sbjct: 684 EKETYRQVLLPQHGY--AVYSPGLYVLSNCICVCTSFLDTRWQLWMMKKYGVAESWTKLM 857
Query: 323 AVPY 326
++P+
Sbjct: 858 SIPH 869
>BQ148450 similar to GP|10177464|dbj gb|AAD21700.1~gene_id:MQB2.18~similar to
unknown protein {Arabidopsis thaliana}, partial (9%)
Length = 666
Score = 117 bits (292), Expect = 1e-26
Identities = 80/195 (41%), Positives = 100/195 (51%), Gaps = 6/195 (3%)
Frame = +3
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLIRNS---ELL 95
++ +EIL RLPVK L+ +CV K S IS P FVK HL +S TR LL + EL+
Sbjct: 12 EIQVEILSRLPVKYLMQFQCVCKLWKSQISKPDFVKKHLRVSNTRHLFLLTFSKLSPELV 191
Query: 96 LVDSRLPSV-TAIIPDTTH-NFRLNPSDNHPIMIDSCDGIICFENRNDNHVDLVVWNPCT 153
+ L SV T + P T + LN D M+ SC GI+C + N V+WNP
Sbjct: 192 IKSYPLSSVFTEMTPTFTQLEYPLNNRDESDSMVGSCHGILCIQC---NLSFPVLWNPSI 362
Query: 154 GKFKILPPLENIPNGKTHTLYSIGYDRFVDNYKVVAFSCHRQINKSYKYCNSQVRVHTLG 213
KF LP E N + Y+ GYD D YKVVA C I+ + V VHT+G
Sbjct: 363 RKFTKLPSFEFPQNKFINPTYAFGYDHSSDTYKVVAVFCTSNIDNGVYQLKTLVNVHTMG 542
Query: 214 TNFWRRI-PNFPSNI 227
TN WRRI FP I
Sbjct: 543 TNCWRRIQTEFPFKI 587
>BG455472
Length = 505
Score = 101 bits (252), Expect(2) = 1e-25
Identities = 60/155 (38%), Positives = 84/155 (53%), Gaps = 1/155 (0%)
Frame = +3
Query: 172 TLYSIGYDRFVDNYKVVAFSCHRQINKSYKYCNSQVRVHTLGTNFWRRIPNFPSNIMGLP 231
T S GYD F+DNYKV+ ++VR +TLGT++WR+I + P
Sbjct: 96 TSLSFGYDHFIDNYKVIVXXDE-----------NEVRXNTLGTDYWRKIEDIHCYTKYGP 242
Query: 232 NGYVGKFVSGTINWAIENQKNYDSW-VILSLDLGNESYQEISRPDFGLDDPVHIFTLGVS 290
G FV GT+NW W VI+SLDL ES QE+ PDFG ++ + + LGV
Sbjct: 243 ----GIFVCGTVNWV--------XWDVIISLDLXKESCQELCPPDFGNEN--NWWNLGVL 380
Query: 291 KDCLCVLVYTETLLGIWVMKDYGNKNSWTKLFAVP 325
+D L V ++ +W+MK+ GNK WTKL+ +P
Sbjct: 381 RDXL*VFAGSDEYWDVWIMKEXGNKEXWTKLYTIP 485
Score = 32.7 bits (73), Expect(2) = 1e-25
Identities = 13/23 (56%), Positives = 16/23 (69%)
Frame = +2
Query: 142 NHVDLVVWNPCTGKFKILPPLEN 164
N+ +WNP KFK+LPPLEN
Sbjct: 17 NNDXYFLWNPSITKFKLLPPLEN 85
>TC81981 similar to GP|11994545|dbj|BAB02732.
emb|CAB62106.1~gene_id:MGD8.12~similar to unknown
protein {Arabidopsis thaliana}, partial (4%)
Length = 635
Score = 95.1 bits (235), Expect(2) = 1e-23
Identities = 71/185 (38%), Positives = 96/185 (51%), Gaps = 14/185 (7%)
Frame = +2
Query: 18 STTVRKQNSSD---PFGLFALLWLDLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVK 74
+TT RK+++ P L DL EIL RLPVK L+ L+C+ + NSLIS+PKF K
Sbjct: 5 TTTHRKRSTGTLTCPPPELPTLPFDLLPEILCRLPVKLLVQLRCLCEFFNSLISNPKFAK 184
Query: 75 HHLHLSQTRPYHLL-----IRNSELLLVDSRLPSVTAIIPDTTH------NFRLNPSDNH 123
HL LS T+ +HLL I E + D +P V + T N NP N
Sbjct: 185 KHLQLS-TKRHHLLVTSWNISRGEFVQHDFPIPLVFSTSTAVTQTQLYAPNILTNPR-NF 358
Query: 124 PIMIDSCDGIICFENRNDNHVDLVVWNPCTGKFKILPPLENIPNGKTHTLYSIGYDRFVD 183
P ++ +GI+C + N ++NP KFK+LPP EN P + GYD F+D
Sbjct: 359 PTVMCCSEGILCGQL---NLGSYFLYNPSIRKFKLLPPFEN-PCEHVPLYINFGYDHFID 526
Query: 184 NYKVV 188
NYKV+
Sbjct: 527 NYKVI 541
Score = 32.7 bits (73), Expect(2) = 1e-23
Identities = 12/23 (52%), Positives = 19/23 (82%)
Frame = +1
Query: 205 SQVRVHTLGTNFWRRIPNFPSNI 227
++V V+TLGT++W+RI + P NI
Sbjct: 559 NEVSVYTLGTDYWKRIEDIPYNI 627
>AW256529 homologue to GP|496669|emb|C C-169 protein {Saccharomyces
cerevisiae}, partial (7%)
Length = 624
Score = 104 bits (260), Expect = 5e-23
Identities = 66/178 (37%), Positives = 97/178 (54%), Gaps = 6/178 (3%)
Frame = +1
Query: 219 RIPNFPSNIMGLPNGYVGKFVSGTINWAIENQ--KNYDSWVILSLDLGNESYQEISRPDF 276
+I +FPS + +P G FVSGT+NW + N I+SLDL E YQEIS+PD+
Sbjct: 1 KIEDFPS--LMIPYSQHGIFVSGTVNWLADYDLDDNNSLGTIVSLDLRKEIYQEISQPDY 174
Query: 277 GLDDPVHIFTLGVSKDCLCVLVYTETLLGIWVMKDYGNKNSWTKLFAVPYAKVGYHGFGF 336
G D +LG ++CLCV ++++ +W+MK+YGN+ SW KL +P F
Sbjct: 175 G--DVTVKLSLGAMRECLCVFSHSDSFDDVWLMKEYGNEESWIKLIRIPCISNRSLVFDN 348
Query: 337 VDLHYISEEDDQVFL----HFCSKVYVYNYKNSTVKTLDIQGQPSILYNSSRVYFESL 390
V++ Y+ E+D V L F K VY+ +N +K+ Q + S+VY ESL
Sbjct: 349 VNILYVFEDDRHVLLLLEERFKLKWVVYDSENGFIKSAKSQ---DFSWVESKVYVESL 513
>TC84654
Length = 605
Score = 101 bits (251), Expect = 6e-22
Identities = 58/134 (43%), Positives = 83/134 (61%)
Frame = +1
Query: 257 VILSLDLGNESYQEISRPDFGLDDPVHIFTLGVSKDCLCVLVYTETLLGIWVMKDYGNKN 316
VI+SLDL NESY+ + PDFG + V TL V DC+C+L ++ T +WVMK++GN+N
Sbjct: 13 VIVSLDLENESYRALLFPDFG-EMNVEALTLEVLMDCMCLLCHSGTFSDVWVMKEFGNEN 189
Query: 317 SWTKLFAVPYAKVGYHGFGFVDLHYISEEDDQVFLHFCSKVYVYNYKNSTVKTLDIQGQP 376
SW +LF VPY + G + Y+ EDDQV L SK+ +YN ++ T K+L+IQ
Sbjct: 190 SWARLFRVPYME-GVGSGPYTKAFYV-YEDDQVLLECQSKLVLYNSRDGTFKSLEIQSTD 363
Query: 377 SILYNSSRVYFESL 390
+ +VY +SL
Sbjct: 364 G--WMVPQVYQQSL 399
>AJ498810 weakly similar to PIR|D86339|D863 protein F2D10.14 [imported] -
Arabidopsis thaliana, partial (6%)
Length = 715
Score = 96.7 bits (239), Expect = 1e-20
Identities = 63/150 (42%), Positives = 85/150 (56%), Gaps = 4/150 (2%)
Frame = +3
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLIRNSELLLVD 98
D+ EIL RLPVK L+ L+C+ K NSLISDP F K HL LS T+ +HL++ N EL++ D
Sbjct: 279 DVLPEILCRLPVKLLIQLRCLCKFFNSLISDPTFAKKHLQLS-TKRHHLVLTNDELVMYD 455
Query: 99 SRLPSV--TAIIPDTTHNFRLNPSDNHPIMID--SCDGIICFENRNDNHVDLVVWNPCTG 154
S +PS+ T+ + T + N I SCDGI + ++ +WNP
Sbjct: 456 SPIPSLFSTSAVFTQTQLHISSTLTNGRIYAPTCSCDGIFLGMLKVGSY---YLWNPSIR 626
Query: 155 KFKILPPLENIPNGKTHTLYSIGYDRFVDN 184
KFK+LP LEN P+ S GYD +DN
Sbjct: 627 KFKLLPTLEN-PHEHGANFCSFGYDHLIDN 713
>TC89815
Length = 807
Score = 96.3 bits (238), Expect = 2e-20
Identities = 74/247 (29%), Positives = 124/247 (49%), Gaps = 14/247 (5%)
Frame = +1
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLIR-NSELLLV 97
++F++IL LP LL + SKSL S+I F +LHL + ++L+IR N+ L +
Sbjct: 55 EIFIDILSLLPPHPLLRFRSTSKSLKSIIDSHTFT--NLHLKNSNNFYLIIRHNANLYQL 228
Query: 98 DSRLPSVTAIIPDTTHNFRLNPSDNHPIMIDSCDGIICFENRNDNHVDLVVWNPCTGKFK 157
D P++T IP N L N + SC+G+IC N D D+ WNP K +
Sbjct: 229 D--FPNLTPPIP---LNHPLMSYSNRITLFGSCNGLICISNIAD---DIAFWNPNIRKHR 384
Query: 158 ILPPLENIPNGKTHT------LYSIGYDRFVDNYKVVAFSCHRQI-NKSYKYCNSQVRVH 210
I+P L P ++ T ++ GYD F +YK+V S + N+S+ +SQVRV
Sbjct: 385 IIPYLPTTPRSESDTTLFAARVHGFGYDSFAGDYKLVRISYFVDLQNRSF---DSQVRVF 555
Query: 211 TLGTNFWRRIPNFP-----SNIMGLPNGYVGKFVSGTINWAIENQ-KNYDSWVILSLDLG 264
+L N W+ +P+ + MG+ S +++W + + + + +I++ +L
Sbjct: 556 SLKMNSWKELPSMNYALCCARTMGVFVEDSNNLNSNSLHWVVTRKLEPFQPDLIVAFNLT 735
Query: 265 NESYQEI 271
E + E+
Sbjct: 736 LEIFNEV 756
>TC81493 weakly similar to PIR|F86249|F86249 protein F25C20.23 [imported] -
Arabidopsis thaliana, partial (9%)
Length = 728
Score = 91.7 bits (226), Expect = 5e-19
Identities = 78/253 (30%), Positives = 124/253 (48%), Gaps = 21/253 (8%)
Frame = +1
Query: 43 EILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLIRNSELLLVDSRLP 102
EIL R+P K LL L+ K +LI F+ LHLS++R +++R SRL
Sbjct: 37 EILSRVPAKPLLRLRSTCKWWRNLIDSTDFI--FLHLSKSRDSVIILRQ------HSRLY 192
Query: 103 SVTAIIPDTTHNFRLNPSDNHPIM--------IDSCDGIICFENRNDNHVDLVVWNPCTG 154
+ D R+ D HP+M + SC+G++C N D D+ WNP
Sbjct: 193 EL-----DLNSMDRVKELD-HPLMCYSNRIKVLGSCNGLLCICNIAD---DIAFWNPTIR 345
Query: 155 KFKILP--PLENIPNGKTHTL--------YSIGYDRFVDNYKVVAFSCHRQI-NKSYKYC 203
K +I+P PL + +T+ Y GYD D+YK+V+ S + N+SY
Sbjct: 346 KHRIIPSEPLIRKETNENNTITTLLAAHVYGFGYDSATDDYKLVSISNFVDLHNRSY--- 516
Query: 204 NSQVRVHTLGTNFWRRIPNFPSNI-MGLPNGYVGKFVSGTINWAIENQKNYDSW-VILSL 261
+S V ++T+G++ W +P P + P +G FVSG ++W + DS +I++
Sbjct: 517 DSHVTIYTMGSDVWMPLPGVPYALCCARP---MGVFVSGALHWVVPRALEPDSRDLIVAF 687
Query: 262 DLGNESYQEISRP 274
DL E ++E++ P
Sbjct: 688 DLRFEVFREVALP 726
>TC84199 similar to GP|9294656|dbj|BAB03005.1
gb|AAF30317.1~gene_id:F14O13.6~similar to unknown
protein {Arabidopsis thaliana}, partial (14%)
Length = 673
Score = 85.9 bits (211), Expect = 3e-17
Identities = 65/195 (33%), Positives = 90/195 (45%), Gaps = 6/195 (3%)
Frame = +1
Query: 4 PLTHSKRKHRHNVVSTTVRKQNSSDPFGLFALLWLDLFLEILYRLPVKSLLVLKCVSKSL 63
P T + R T +K+ + P+ L L+L ++IL LPVKSLL KCV KS
Sbjct: 28 PQTLTMRNSCREKRRTVKKKEEKTAPY-----LPLELIIQILLWLPVKSLLRFKCVCKSW 192
Query: 64 NSLISDPKFVKHHLHLSQTRPYHLLIRNSEL---LLVD---SRLPSVTAIIPDTTHNFRL 117
SLISD F H ++ +L + + L +D + + IP+ NF
Sbjct: 193 FSLISDTHFANSHFQITAKHSRRVLFMLNHVPTTLSLDFEALHCDNAVSEIPNPIPNFVE 372
Query: 118 NPSDNHPIMIDSCDGIICFENRNDNHVDLVVWNPCTGKFKILPPLENIPNGKTHTLYSIG 177
P D+ SC G I N DL +WNP T +K +P N N H LY G
Sbjct: 373 PPCDSLDTNSSSCRGFIFLH----NDPDLFIWNPSTRVYKQIPLSPNDSN-SFHCLYGFG 537
Query: 178 YDRFVDNYKVVAFSC 192
YD+ D+Y VV+ +C
Sbjct: 538 YDQLRDDYLVVSVTC 582
>TC84657 weakly similar to GP|9294656|dbj|BAB03005.1
gb|AAF30317.1~gene_id:F14O13.6~similar to unknown
protein {Arabidopsis thaliana}, partial (14%)
Length = 884
Score = 85.9 bits (211), Expect = 3e-17
Identities = 59/185 (31%), Positives = 91/185 (48%), Gaps = 6/185 (3%)
Frame = +1
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLIRNSELLLVD 98
+L ++I+ RLPVKSL+ KCV KSL +LISD F K H LS + ++ S L L
Sbjct: 337 ELIIQIMLRLPVKSLIRFKCVCKSLLALISDHNFAKSHFELSTATHTNRIVFMSTLALET 516
Query: 99 SRLPSVTAIIPD---TTHNFRLNPSDNHPIM--IDSCDGIICFENRNDNHVDLVVWNPCT 153
+ ++ D T+ N P +++ + SC G I + ++ +WNP T
Sbjct: 517 RSIDFEASLNDDSASTSLNLNFMPPESYSSLEIKSSCRGFIVLTCSS----NIYLWNPST 684
Query: 154 GKFKILP-PLENIPNGKTHTLYSIGYDRFVDNYKVVAFSCHRQINKSYKYCNSQVRVHTL 212
G K +P P N+ + LY GYD D+Y VV+ S + I+ +S ++ +L
Sbjct: 685 GHHKQIPFPASNLDAKYSCCLYGFGYDHLRDDYLVVSVSYNTSIDPVDDNISSHLKFFSL 864
Query: 213 GTNFW 217
N W
Sbjct: 865 RANTW 879
>BG646118
Length = 764
Score = 78.2 bits (191), Expect = 5e-15
Identities = 62/195 (31%), Positives = 100/195 (50%), Gaps = 11/195 (5%)
Frame = -1
Query: 221 PNFPSNIMGLPNGYVGKFVSGTINWAIENQKNYDSWV---ILSLDLGNESYQEISRPDFG 277
P FP + P ++GK VSGTINW + Q YD+ I+SLDLG+ES +E+ P
Sbjct: 752 PKFPFGCV--PLQFLGKSVSGTINWLVARQ--YDNKFQDCIVSLDLGSESCKEVLLPKEV 585
Query: 278 LDDPVHI-FTLGVSKDCLCVLVYTETLLGIWVMKDYGNKNSWTKLFAVPYAKVGYHGFGF 336
+ + + LGV +DCLC++ + +W+MK++GN +SW KL++V + + F
Sbjct: 584 DTSTLRLRWYLGVLRDCLCLVFGHD----VWIMKEHGNNDSWAKLYSVSFMRDFPSSFAT 417
Query: 337 VDLHYISEEDDQVFLHFC-------SKVYVYNYKNSTVKTLDIQGQPSILYNSSRVYFES 389
+++ +I + D L C K+ +N +N T K ++ S++ +
Sbjct: 416 IEVLHIFK--DGCLLLDCIYEGERTRKLVFHNSRNGTFK-----------FSESKIMRD- 279
Query: 390 LYDSSGVYVESLISP 404
V VESLISP
Sbjct: 278 ------VCVESLISP 252
>TC91177 similar to GP|9294656|dbj|BAB03005.1
gb|AAF30317.1~gene_id:F14O13.6~similar to unknown
protein {Arabidopsis thaliana}, partial (9%)
Length = 1298
Score = 72.0 bits (175), Expect(2) = 1e-14
Identities = 78/304 (25%), Positives = 121/304 (39%), Gaps = 26/304 (8%)
Frame = +2
Query: 57 KCVSKSLNSLISDPKFVKHHLHLSQTR---------PY-HLLIRNSELLLVDSRLPSVTA 106
KCV K SLIS P F H L+ PY L + EL L D T
Sbjct: 77 KCVCKLWLSLISQPHFANSHFQLTTATHTNRIMLITPYLQSLSIDLELSLNDDSAVYTTD 256
Query: 107 I--IPDTTHNFRLNPSDNHPI-------MID---SCDGIICFENRNDNHVDLVVWNPCTG 154
I + D + + SD + ++D SC G I + + L +WNP TG
Sbjct: 257 ISFLIDDEDYYSSSSSDMDDLSPPKSFFILDFKGSCRGFILL----NCYSSLCIWNPSTG 424
Query: 155 KFKILPPLENIPNGKTHTLYSIGYDRFVDNYKVVAFSCHRQINKSYKYCNSQVRVHTLGT 214
K +P N + Y GYD D+Y V++ S + + S S + + +L
Sbjct: 425 FHKRIPFTTIDSNPDANYFYGFGYDESTDDYLVISMS--YEPSPSSDGMLSHLGIFSLRA 598
Query: 215 NFWRRIP--NFPSNIMGLPNGYVGKFVSGTINWAIENQKNYDSWVILSLDLGNESYQEIS 272
N W R+ N V +G I+W + + + VI++ L E+
Sbjct: 599 NVWTRVEGGNLLLYSQNSLLNLVESLSNGAIHW-LAFRNDISMPVIVAFHLMERKLLELR 775
Query: 273 RPDFGLDDPVHIFTLGVSKDCLCV--LVYTETLLGIWVMKDYGNKNSWTKLFAVPYAKVG 330
P+ ++ P + L V + CL + ++ IWVM+ Y ++SWTK + +
Sbjct: 776 LPNEIINGPSRAYDLWVYRGCLALWHILPDRVTFQIWVMEKYNVQSSWTKTLVLSFDGNP 955
Query: 331 YHGF 334
H F
Sbjct: 956 AHSF 967
Score = 25.0 bits (53), Expect(2) = 1e-14
Identities = 11/18 (61%), Positives = 15/18 (83%)
Frame = +3
Query: 39 DLFLEILYRLPVKSLLVL 56
+L ++IL RLPVKSL+ L
Sbjct: 24 ELIIQILLRLPVKSLIRL 77
>BG649101 homologue to GP|6648199|gb|A unknown protein {Arabidopsis
thaliana}, partial (3%)
Length = 799
Score = 76.3 bits (186), Expect = 2e-14
Identities = 74/229 (32%), Positives = 103/229 (44%), Gaps = 22/229 (9%)
Frame = +1
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLIR-------N 91
DL IL LPVK++ LK VSKS N+LI+ P F+K HL+ S P +L N
Sbjct: 94 DLIAIILTFLPVKTITQLKLVSKSWNTLITSPSFIKIHLNQSSQNPNFILTPSRKQYSIN 273
Query: 92 SELLLVDSRLPSVTAIIPDTTHNFRLNPSDNHPIMIDSCDGIIC--FENRNDNHVD--LV 147
+ L + RL + + DT HN N D+H ++ SC+G++C F++ H+
Sbjct: 274 NVLSVPIPRLLTGNTVSGDTYHNILNN--DHHFRVVGSCNGLLCLLFKSEFITHLKFRFR 447
Query: 148 VWNPCT-------GKFKILPPLENIPNGKTHTLYSIGYDRFVDNYKVVAFSCHRQINKSY 200
+WNP T G F+ PL + ++ G D YK+VA +
Sbjct: 448 IWNPATRTISEELGFFRKYKPLFG-----GVSRFTFGCDYLRGTYKLVAL----HTVEDG 600
Query: 201 KYCNSQVRVHTLGTN----FWRRIPNFPSNIMGLPNGYVGKFVSGTINW 245
S VRV LG + WR IPN P G +SGT NW
Sbjct: 601 DVMRSNVRVFNLGNDDSDKCWRNIPN-PFVCAD------GVHLSGTGNW 726
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.140 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,162,273
Number of Sequences: 36976
Number of extensions: 281902
Number of successful extensions: 1719
Number of sequences better than 10.0: 135
Number of HSP's better than 10.0 without gapping: 1611
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1634
length of query: 404
length of database: 9,014,727
effective HSP length: 98
effective length of query: 306
effective length of database: 5,391,079
effective search space: 1649670174
effective search space used: 1649670174
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC122729.3


