

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122724.2 + phase: 0
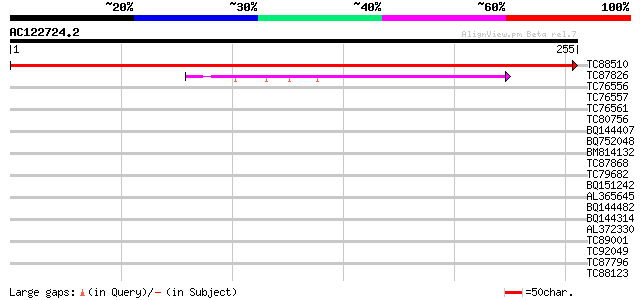
(255 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88510 similar to GP|12849485|dbj|BAB28361. data source:MGD so... 514 e-147
TC87826 similar to SP|Q03211|EXLP_TOBAC Pistil-specific extensin... 97 6e-21
TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 38 0.003
TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 38 0.003
TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 38 0.003
TC80756 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 37 0.010
BQ144407 weakly similar to GP|6523547|emb| hydroxyproline-rich g... 36 0.013
BQ752048 weakly similar to GP|7303636|gb|A CG13214-PA {Drosophil... 36 0.013
BM814132 weakly similar to OMNI|MT3615.1 PE_PGRS family protein ... 36 0.013
TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 35 0.021
TC79682 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproli... 35 0.028
BQ151242 similar to PIR|G86292|G86 hypothetical protein AAF82153... 34 0.048
AL365645 34 0.048
BQ144482 similar to GP|6523547|emb| hydroxyproline-rich glycopro... 34 0.063
BQ144314 weakly similar to GP|21322711|em pherophorin-dz1 protei... 34 0.063
AL372330 similar to GP|21322752|dbj cold shock protein-1 {Tritic... 34 0.063
TC89001 similar to GP|5726567|gb|AAD48471.1| glycine-rich RNA-bi... 33 0.082
TC92049 similar to PIR|T01061|T01061 hypothetical protein YUP8H1... 33 0.11
TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear prot... 33 0.11
TC88123 similar to GP|8777484|dbj|BAA97064.1 contains similarity... 33 0.11
>TC88510 similar to GP|12849485|dbj|BAB28361. data source:MGD source
key:MGI:95486 evidence:ISS~fibrillarin~putative {Mus
musculus}, partial (5%)
Length = 1141
Score = 514 bits (1325), Expect = e-147
Identities = 255/255 (100%), Positives = 255/255 (100%)
Frame = +3
Query: 1 MNAIQVAASNPAIYVRKNYQPHHFLSTPKGLSMNANPLSRTFLIPRAKLSSGLYCVRQSA 60
MNAIQVAASNPAIYVRKNYQPHHFLSTPKGLSMNANPLSRTFLIPRAKLSSGLYCVRQSA
Sbjct: 99 MNAIQVAASNPAIYVRKNYQPHHFLSTPKGLSMNANPLSRTFLIPRAKLSSGLYCVRQSA 278
Query: 61 TTAPVLKVYPSNLTQSVPVLKSQKPRHVCLAGGKGMMDNSEDSQRKSLEEAMKSLQEKIQ 120
TTAPVLKVYPSNLTQSVPVLKSQKPRHVCLAGGKGMMDNSEDSQRKSLEEAMKSLQEKIQ
Sbjct: 279 TTAPVLKVYPSNLTQSVPVLKSQKPRHVCLAGGKGMMDNSEDSQRKSLEEAMKSLQEKIQ 458
Query: 121 KGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMSDETAQIVFATIGFMFVYIYVING 180
KGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMSDETAQIVFATIGFMFVYIYVING
Sbjct: 459 KGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMSDETAQIVFATIGFMFVYIYVING 638
Query: 181 VELTKLARDFIKYLLGGTQSVRLKRASYKCSRFYKKITGQNEVDEDGLETGPLRWNIADF 240
VELTKLARDFIKYLLGGTQSVRLKRASYKCSRFYKKITGQNEVDEDGLETGPLRWNIADF
Sbjct: 639 VELTKLARDFIKYLLGGTQSVRLKRASYKCSRFYKKITGQNEVDEDGLETGPLRWNIADF 818
Query: 241 YRDVLRNYMKPNSNE 255
YRDVLRNYMKPNSNE
Sbjct: 819 YRDVLRNYMKPNSNE 863
>TC87826 similar to SP|Q03211|EXLP_TOBAC Pistil-specific extensin-like
protein precursor (PELP). [Common tobacco] {Nicotiana
tabacum}, partial (6%)
Length = 1222
Score = 97.1 bits (240), Expect = 6e-21
Identities = 63/159 (39%), Positives = 90/159 (55%), Gaps = 13/159 (8%)
Frame = -3
Query: 80 LKSQKPRHVCLAGGKGMMDNS-EDSQRKSLEEAMKS---------LQEKIQKGEY--SGG 127
LKS KP AGGKGM +NS E+S +S+ AM L+++IQKGEY +GG
Sbjct: 821 LKSLKPLR---AGGKGMSENSDENSPWQSITNAMDKFKGQPIEEVLRKQIQKGEYLDNGG 651
Query: 128 SGSKPPGGRG-RGGGGGNSDGSEEGSTGGMSDETAQIVFATIGFMFVYIYVINGVELTKL 186
SG KPPGG G GG GG + GS S +T Q +FA+ + +YIY+I G EL +
Sbjct: 650 SGVKPPGGGGGEGGSGGGNPNGPGGSDDEESYDTRQTIFASGALLSLYIYLIMGKELLRT 471
Query: 187 ARDFIKYLLGGTQSVRLKRASYKCSRFYKKITGQNEVDE 225
DF+++++G ++ R+KR + + YK + DE
Sbjct: 470 IFDFLRFIVGLGKTSRIKRILVRLGKLYKSTKRRKITDE 354
>TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 754
Score = 38.1 bits (87), Expect = 0.003
Identities = 23/68 (33%), Positives = 30/68 (43%)
Frame = +2
Query: 88 VCLAGGKGMMDNSEDSQRKSLEEAMKSLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDG 147
V A K M D E + L+ ++ + +G GG G + GG G GGGGG G
Sbjct: 239 VTFANEKSMNDAIEAMNGQDLDGRNITVNQAQSRGSGGGGGGGR--GGGGYGGGGGGYGG 412
Query: 148 SEEGSTGG 155
G GG
Sbjct: 413 ERRGYGGG 436
Score = 32.0 bits (71), Expect = 0.24
Identities = 18/36 (50%), Positives = 18/36 (50%), Gaps = 4/36 (11%)
Frame = +2
Query: 124 YSGGSGSKPPGGRG----RGGGGGNSDGSEEGSTGG 155
Y GG G GG G RGGGGG S G G GG
Sbjct: 425 YGGGGGYGGGGGGGYGERRGGGGGYSRGGGGGGYGG 532
Score = 28.9 bits (63), Expect = 2.0
Identities = 21/55 (38%), Positives = 26/55 (47%), Gaps = 7/55 (12%)
Frame = +2
Query: 122 GEYSGGSGSKPPGGRG--RGGGGGNSDG---SEEGSTGG--MSDETAQIVFATIG 169
G GG G + GG G RGGGGG G S G GG + D + I+ +G
Sbjct: 449 GGGGGGYGERRGGGGGYSRGGGGGGYGGGGYSRGGGDGGY*VLDLSFMIIILDVG 613
>TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 912
Score = 38.1 bits (87), Expect = 0.003
Identities = 23/68 (33%), Positives = 30/68 (43%)
Frame = +1
Query: 88 VCLAGGKGMMDNSEDSQRKSLEEAMKSLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDG 147
V A K M D E + L+ ++ + +G GG G + GG G GGGGG G
Sbjct: 397 VTFANEKSMNDAIEAMNGQDLDGRNITVNQAQSRGSGGGGGGGR--GGGGYGGGGGGYGG 570
Query: 148 SEEGSTGG 155
G GG
Sbjct: 571 ERRGYGGG 594
Score = 32.0 bits (71), Expect = 0.24
Identities = 18/36 (50%), Positives = 18/36 (50%), Gaps = 4/36 (11%)
Frame = +1
Query: 124 YSGGSGSKPPGGRG----RGGGGGNSDGSEEGSTGG 155
Y GG G GG G RGGGGG S G G GG
Sbjct: 583 YGGGGGYGGGGGGGYGERRGGGGGYSRGGGGGGYGG 690
Score = 28.9 bits (63), Expect = 2.0
Identities = 21/55 (38%), Positives = 26/55 (47%), Gaps = 7/55 (12%)
Frame = +1
Query: 122 GEYSGGSGSKPPGGRG--RGGGGGNSDG---SEEGSTGG--MSDETAQIVFATIG 169
G GG G + GG G RGGGGG G S G GG + D + I+ +G
Sbjct: 607 GGGGGGYGERRGGGGGYSRGGGGGGYGGGGYSRGGGDGGY*VLDLSFMIIILDVG 771
>TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 1054
Score = 38.1 bits (87), Expect = 0.003
Identities = 23/68 (33%), Positives = 30/68 (43%)
Frame = +1
Query: 88 VCLAGGKGMMDNSEDSQRKSLEEAMKSLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDG 147
V A K M D E + L+ ++ + +G GG G + GG G GGGGG G
Sbjct: 493 VTFANEKSMNDAIEAMNGQDLDGRNITVNQAQSRGSGGGGGGGR--GGGGYGGGGGGYGG 666
Query: 148 SEEGSTGG 155
G GG
Sbjct: 667 ERRGYGGG 690
Score = 32.0 bits (71), Expect = 0.24
Identities = 18/36 (50%), Positives = 18/36 (50%), Gaps = 4/36 (11%)
Frame = +1
Query: 124 YSGGSGSKPPGGRG----RGGGGGNSDGSEEGSTGG 155
Y GG G GG G RGGGGG S G G GG
Sbjct: 679 YGGGGGYGGGGGGGYGERRGGGGGYSRGGGGGGYGG 786
Score = 28.9 bits (63), Expect = 2.0
Identities = 21/55 (38%), Positives = 26/55 (47%), Gaps = 7/55 (12%)
Frame = +1
Query: 122 GEYSGGSGSKPPGGRG--RGGGGGNSDG---SEEGSTGG--MSDETAQIVFATIG 169
G GG G + GG G RGGGGG G S G GG + D + I+ +G
Sbjct: 703 GGGGGGYGERRGGGGGYSRGGGGGGYGGGGYSRGGGDGGY*VLDLSFMIIILDVG 867
>TC80756 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (53%)
Length = 1189
Score = 36.6 bits (83), Expect = 0.010
Identities = 29/73 (39%), Positives = 36/73 (48%), Gaps = 1/73 (1%)
Frame = +3
Query: 97 MDNSEDSQRKSLEEAMKSLQEKIQKGEYSGGSGSKPPGGRGR-GGGGGNSDGSEEGSTGG 155
+D ED Q K LE + K+ Y GG+G P G G GGGGG S+G+ GS G
Sbjct: 879 VDEFEDKQ-KELEGICNPIIAKM----YQGGAGGDVPMGDGMPGGGGGASNGT--GSGAG 1037
Query: 156 MSDETAQIVFATI 168
E VF T+
Sbjct: 1038PKIEEVD*VFETL 1076
>BQ144407 weakly similar to GP|6523547|emb| hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (23%)
Length = 1217
Score = 36.2 bits (82), Expect = 0.013
Identities = 17/31 (54%), Positives = 19/31 (60%)
Frame = +1
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGS 152
GE GG G GG GRGGGGG +G E G+
Sbjct: 157 GEGWGGRGRPGGGGGGRGGGGGRRNGGEIGA 249
Score = 28.1 bits (61), Expect = 3.4
Identities = 15/34 (44%), Positives = 16/34 (46%), Gaps = 2/34 (5%)
Frame = +2
Query: 127 GSGSKPPGGRGRGGG--GGNSDGSEEGSTGGMSD 158
G G+ GG G GGG GG G G G SD
Sbjct: 167 GGGAAGRGGAGGGGGVGGGEETGGRSGPWTGSSD 268
Score = 28.1 bits (61), Expect = 3.4
Identities = 14/39 (35%), Positives = 20/39 (50%)
Frame = +3
Query: 123 EYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMSDETA 161
++ G G++P GG GRGG GG + G G+ A
Sbjct: 153 KWRGMGGARPAGG-GRGGAGGWGGEKKRGGDRGLGPGAA 266
>BQ752048 weakly similar to GP|7303636|gb|A CG13214-PA {Drosophila
melanogaster}, partial (6%)
Length = 657
Score = 36.2 bits (82), Expect = 0.013
Identities = 19/49 (38%), Positives = 24/49 (48%), Gaps = 1/49 (2%)
Frame = -3
Query: 108 LEEAMKSLQEKI-QKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
L EA + + ++ + Y GG G GG GRG GGG G G GG
Sbjct: 646 LREAHQEIDPRLLEMTRYGGGGGGGRYGGWGRGRGGGRGGGRGHGGGGG 500
Score = 30.8 bits (68), Expect = 0.53
Identities = 14/31 (45%), Positives = 18/31 (57%), Gaps = 3/31 (9%)
Frame = -3
Query: 122 GEYSG---GSGSKPPGGRGRGGGGGNSDGSE 149
G Y G G G GGRG GGGGG ++ ++
Sbjct: 574 GRYGGWGRGRGGGRGGGRGHGGGGGGNNAND 482
>BM814132 weakly similar to OMNI|MT3615.1 PE_PGRS family protein
{Mycobacterium tuberculosis CDC1551}, partial (7%)
Length = 164
Score = 36.2 bits (82), Expect = 0.013
Identities = 17/34 (50%), Positives = 17/34 (50%)
Frame = +1
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G GG G GG GRGGGGG G G GG
Sbjct: 16 GGGGGGGGGGGGGGGGRGGGGGGGGGGAGGGGGG 117
Score = 35.8 bits (81), Expect = 0.016
Identities = 17/34 (50%), Positives = 17/34 (50%)
Frame = +2
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G GG G GG GRGGGGG G G GG
Sbjct: 50 GGGGGGGGGGGGGGGGRGGGGGGGGGGGGGGGGG 151
Score = 35.8 bits (81), Expect = 0.016
Identities = 17/34 (50%), Positives = 17/34 (50%)
Frame = +2
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G GG G GGRG GGGGG G G GG
Sbjct: 56 GGGGGGGGGGGGGGRGGGGGGGGGGGGGGGGGGG 157
Score = 33.5 bits (75), Expect = 0.082
Identities = 16/34 (47%), Positives = 16/34 (47%)
Frame = +1
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G GG G GGRG GGGGG G GG
Sbjct: 22 GGGGGGGGGGGGGGRGGGGGGGGGGAGGGGGGGG 123
Score = 33.5 bits (75), Expect = 0.082
Identities = 16/34 (47%), Positives = 17/34 (49%)
Frame = +3
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G +GG G GG G GGGGG G G GG
Sbjct: 54 GGGAGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 155
Score = 33.5 bits (75), Expect = 0.082
Identities = 16/34 (47%), Positives = 17/34 (49%)
Frame = +3
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G GG+G GG G GGGGG G G GG
Sbjct: 45 GGGGGGAGGGGGGGGGGGGGGGGGGGGGGGGGGG 146
Score = 33.1 bits (74), Expect = 0.11
Identities = 15/30 (50%), Positives = 16/30 (53%)
Frame = +2
Query: 126 GGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
GG+G GG G GGGGG G G GG
Sbjct: 5 GGAGGGGGGGGGGGGGGGGGGGGGGGGGGG 94
Score = 30.4 bits (67), Expect = 0.69
Identities = 15/34 (44%), Positives = 16/34 (46%)
Frame = +2
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G +GG G GG G GGGGG G G G
Sbjct: 2 GGGAGGGGGGGGGGGGGGGGGGGGGGGGGGGGRG 103
>TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana, partial (91%)
Length = 860
Score = 35.4 bits (80), Expect = 0.021
Identities = 18/45 (40%), Positives = 24/45 (53%), Gaps = 6/45 (13%)
Frame = +3
Query: 105 RKSLEEAMKSLQEK------IQKGEYSGGSGSKPPGGRGRGGGGG 143
R+ ++A++ L K + SGG G GGRGRGGGGG
Sbjct: 183 RRDAQDAIRDLDGKNGWRVELSHNSRSGGGGGGGGGGRGRGGGGG 317
>TC79682 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproline-rich
glycoprotein DZ-HRGP {Volvox carteri f. nagariensis},
partial (17%)
Length = 1031
Score = 35.0 bits (79), Expect = 0.028
Identities = 18/37 (48%), Positives = 21/37 (56%)
Frame = -1
Query: 119 IQKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
++ G SGG GSK G GGGGG DG + G T G
Sbjct: 236 LRGGGNSGGKGSKGAGKIDFGGGGGGGDGVKPGITIG 126
>BQ151242 similar to PIR|G86292|G86 hypothetical protein AAF82153.1
[imported] - Arabidopsis thaliana, partial (5%)
Length = 1371
Score = 34.3 bits (77), Expect = 0.048
Identities = 22/71 (30%), Positives = 34/71 (46%)
Frame = +2
Query: 92 GGKGMMDNSEDSQRKSLEEAMKSLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEG 151
GG+G ++ ++K + K+ + + KG G GS+ GGR RG GGG G
Sbjct: 26 GGRG--ESLGRKKKKKKKFFPKTQKRRRGKGGNKKGRGSEGAGGRERGKGGGGGGERGGG 199
Query: 152 STGGMSDETAQ 162
G +E A+
Sbjct: 200 QKGRRKEEGAE 232
Score = 30.4 bits (67), Expect = 0.69
Identities = 21/68 (30%), Positives = 29/68 (41%), Gaps = 4/68 (5%)
Frame = +1
Query: 92 GGKGMMDNSEDSQRK----SLEEAMKSLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDG 147
GG+ + + ++K S + K Q Q+GE G G + G GRGGGG
Sbjct: 22 GGRKRRELGKKKKKKKKIFSQDAKAKKRQGGEQEGERERGRGREREGKGGRGGGGEGRRT 201
Query: 148 SEEGSTGG 155
E GG
Sbjct: 202 KGEKKRGG 225
>AL365645
Length = 552
Score = 34.3 bits (77), Expect = 0.048
Identities = 25/55 (45%), Positives = 33/55 (59%), Gaps = 10/55 (18%)
Frame = -2
Query: 80 LKSQKPRHVCLAGGKGMMDNS-EDSQRKSLEEAM---------KSLQEKIQKGEY 124
LKS KP AGGKGM +NS E+S +S+ AM + L+++IQKGEY
Sbjct: 164 LKSLKPLR---AGGKGMSENSDENSPWQSITNAMDKFKGQPIEEVLRKQIQKGEY 9
>BQ144482 similar to GP|6523547|emb| hydroxyproline-rich glycoprotein DZ-HRGP
{Volvox carteri f. nagariensis}, partial (16%)
Length = 993
Score = 33.9 bits (76), Expect = 0.063
Identities = 19/43 (44%), Positives = 22/43 (50%), Gaps = 9/43 (20%)
Frame = -1
Query: 122 GEYSGGSGSKPPGGRGRGGGG---------GNSDGSEEGSTGG 155
G++SGG+G P GG GRGGG G G EG GG
Sbjct: 393 GQHSGGTG--PGGGPGRGGGARGAGGRGGEGRGQGEAEGGGGG 271
Score = 33.5 bits (75), Expect = 0.082
Identities = 16/48 (33%), Positives = 22/48 (45%)
Frame = -2
Query: 111 AMKSLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMSD 158
A +E+++ G GG G + G GRGGGG +G G D
Sbjct: 314 AKAGAKERLRGGGGGGGGGGRKRGRGGRGGGGAEIEGEGGGKRWARGD 171
Score = 33.1 bits (74), Expect = 0.11
Identities = 17/41 (41%), Positives = 20/41 (48%), Gaps = 7/41 (17%)
Frame = -3
Query: 122 GEYSGGSGSKPPGGRG-------RGGGGGNSDGSEEGSTGG 155
G GG+G + PGGR GGGGG +G E G G
Sbjct: 355 GAGGGGAGGRGPGGRRPGPRRG*GGGGGGEGEGGERGGGEG 233
Score = 30.0 bits (66), Expect = 0.90
Identities = 18/51 (35%), Positives = 20/51 (38%), Gaps = 9/51 (17%)
Frame = -2
Query: 122 GEYSGGSGSKPPGGRG---------RGGGGGNSDGSEEGSTGGMSDETAQI 163
G GG G PG G RGGGGG G + GG A+I
Sbjct: 362 GGRGGGGGRGGPGAGGAKAGAKERLRGGGGGGGGGGRKRGRGGRGGGGAEI 210
Score = 27.7 bits (60), Expect = 4.5
Identities = 14/36 (38%), Positives = 21/36 (57%), Gaps = 5/36 (13%)
Frame = -1
Query: 121 KGEYSGGSGSKPPGGRGR-----GGGGGNSDGSEEG 151
+G + G+G + GRG+ GGGGG G++EG
Sbjct: 351 RGGGARGAGGRGGEGRGQGEAEGGGGGGRGRGAKEG 244
>BQ144314 weakly similar to GP|21322711|em pherophorin-dz1 protein {Volvox
carteri f. nagariensis}, partial (6%)
Length = 1154
Score = 33.9 bits (76), Expect = 0.063
Identities = 15/26 (57%), Positives = 17/26 (64%)
Frame = -2
Query: 126 GGSGSKPPGGRGRGGGGGNSDGSEEG 151
GG GS GG GRGGGGG G+ +G
Sbjct: 85 GGVGSLGGGGGGRGGGGGRKGGNGKG 8
>AL372330 similar to GP|21322752|dbj cold shock protein-1 {Triticum
aestivum}, partial (33%)
Length = 435
Score = 33.9 bits (76), Expect = 0.063
Identities = 16/34 (47%), Positives = 17/34 (49%)
Frame = +2
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G Y G S S P GG GG S G GS+GG
Sbjct: 47 GGYGGSSSSSYPASSSGGGYGGGSSGGYGGSSGG 148
Score = 32.0 bits (71), Expect = 0.24
Identities = 15/34 (44%), Positives = 16/34 (46%)
Frame = +2
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G Y G SG GG G G GG G G +GG
Sbjct: 143 GGYGGSSGGYGGGGYGGGASGGYGGGGYGGGSGG 244
Score = 29.6 bits (65), Expect = 1.2
Identities = 18/35 (51%), Positives = 18/35 (51%), Gaps = 1/35 (2%)
Frame = +2
Query: 122 GEYSGGSGSKPPGGRGRGG-GGGNSDGSEEGSTGG 155
G Y GGSG GG G GG GGG G G GG
Sbjct: 218 GGYGGGSG----GGYGGGGYGGGGYGGGGGGGYGG 310
Score = 28.9 bits (63), Expect = 2.0
Identities = 17/36 (47%), Positives = 17/36 (47%)
Frame = +2
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMS 157
G SGG G G G G GGG S G G GG S
Sbjct: 131 GGSSGGYGGSSGGYGGGGYGGGASGGYGGGGYGGGS 238
>TC89001 similar to GP|5726567|gb|AAD48471.1| glycine-rich RNA-binding
protein {Glycine max}, partial (90%)
Length = 488
Score = 33.5 bits (75), Expect = 0.082
Identities = 23/71 (32%), Positives = 30/71 (41%)
Frame = +3
Query: 88 VCLAGGKGMMDNSEDSQRKSLEEAMKSLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDG 147
V A K M D E+ + ++ ++ E +G SGG G GG GGGG G
Sbjct: 189 VTFAEEKSMRDAIEEMNGQDIDGRNITVNEAQSRG--SGGGGRGGGGGGYGGGGGYGGGG 362
Query: 148 SEEGSTGGMSD 158
G GG D
Sbjct: 363 GGYGGGGGRRD 395
Score = 33.5 bits (75), Expect = 0.082
Identities = 15/34 (44%), Positives = 17/34 (49%)
Frame = +3
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G Y GG G + G GGGGG G + G GG
Sbjct: 363 GGYGGGGGRRDGGYSRSGGGGGYGGGGDRGYGGG 464
Score = 28.1 bits (61), Expect = 3.4
Identities = 14/34 (41%), Positives = 14/34 (41%)
Frame = +3
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G Y GG G GG GGGG G GG
Sbjct: 324 GGYGGGGGYGGGGGGYGGGGGRRDGGYSRSGGGG 425
Score = 26.9 bits (58), Expect = 7.6
Identities = 14/24 (58%), Positives = 14/24 (58%), Gaps = 1/24 (4%)
Frame = +3
Query: 125 SGGSGSKPPGG-RGRGGGGGNSDG 147
SGG G GG RG GGGGG G
Sbjct: 411 SGGGGGYGGGGDRGYGGGGGGGYG 482
>TC92049 similar to PIR|T01061|T01061 hypothetical protein YUP8H12R.45 -
Arabidopsis thaliana, partial (16%)
Length = 1079
Score = 33.1 bits (74), Expect = 0.11
Identities = 24/89 (26%), Positives = 41/89 (45%)
Frame = +1
Query: 26 STPKGLSMNANPLSRTFLIPRAKLSSGLYCVRQSATTAPVLKVYPSNLTQSVPVLKSQKP 85
+TP G+S A ++I + K+SS R + T+P+L +NLT +Q
Sbjct: 490 NTPSGISKGAPDFCTVYVISKGKISSVRNASRAAPHTSPLLNHIHNNLTSED---VNQTA 660
Query: 86 RHVCLAGGKGMMDNSEDSQRKSLEEAMKS 114
+ +C + M D + +E+MKS
Sbjct: 661 QQIC-SRRMNMRDRTSMKPNSWQDESMKS 744
>TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear protein.
[strain B95-8 Human herpesvirus 4] {Epstein-barr
virus}, partial (23%)
Length = 431
Score = 33.1 bits (74), Expect = 0.11
Identities = 17/34 (50%), Positives = 19/34 (55%)
Frame = +1
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G Y GGS GG G GGGGG G+ G+ GG
Sbjct: 112 GGYGGGSPGGGGGGGGSGGGGGGG-GAHGGAYGG 210
Score = 30.8 bits (68), Expect = 0.53
Identities = 16/34 (47%), Positives = 17/34 (49%)
Frame = +1
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G GG G PGG G GGGG G G+ GG
Sbjct: 100 GGTGGGYGGGSPGGGG-GGGGSGGGGGGGGAHGG 198
Score = 26.9 bits (58), Expect = 7.6
Identities = 16/36 (44%), Positives = 17/36 (46%), Gaps = 2/36 (5%)
Frame = +1
Query: 122 GEYSGGSGSKPPGGRGRGG--GGGNSDGSEEGSTGG 155
G GG+G G G GG GGG GS G GG
Sbjct: 46 GGGGGGAGGAHGVGYGSGGGTGGGYGGGSPGGGGGG 153
Score = 26.6 bits (57), Expect = 10.0
Identities = 14/34 (41%), Positives = 14/34 (41%)
Frame = +1
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G SGG GG GGGGG G GG
Sbjct: 85 GYGSGGGTGGGYGGGSPGGGGGGGGSGGGGGGGG 186
>TC88123 similar to GP|8777484|dbj|BAA97064.1 contains similarity to
RNA-binding protein~gene_id:K15M2.15 {Arabidopsis
thaliana}, partial (11%)
Length = 815
Score = 33.1 bits (74), Expect = 0.11
Identities = 16/33 (48%), Positives = 16/33 (48%)
Frame = +1
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTG 154
G Y G SG G G GGGGG G G TG
Sbjct: 85 GHYGGYSGPTAAGYGGSGGGGGGGYGGGSGVTG 183
Score = 29.3 bits (64), Expect = 1.5
Identities = 15/27 (55%), Positives = 17/27 (62%)
Frame = +1
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGS 148
G Y GGSG P G G GGG G++ GS
Sbjct: 151 GGYGGGSGVTGP-GVGAGGGHGSAGGS 228
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.133 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,030,561
Number of Sequences: 36976
Number of extensions: 90718
Number of successful extensions: 1418
Number of sequences better than 10.0: 172
Number of HSP's better than 10.0 without gapping: 973
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1288
length of query: 255
length of database: 9,014,727
effective HSP length: 94
effective length of query: 161
effective length of database: 5,538,983
effective search space: 891776263
effective search space used: 891776263
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC122724.2


