

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122723.7 + phase: 0 /pseudo
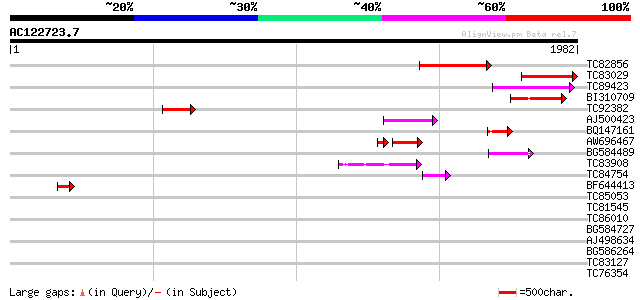
(1982 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC82856 homologue to PIR|T49914|T49914 callose synthase catalyti... 496 e-140
TC83029 similar to GP|20330757|gb|AAM19120.1 Putative glucan syn... 382 e-105
TC89423 similar to GP|20330757|gb|AAM19120.1 Putative glucan syn... 185 2e-46
BI310709 similar to GP|20330757|g Putative glucan synthase {Oryz... 178 2e-44
TC92382 similar to PIR|E86189|E86189 hypothetical protein [impor... 173 7e-43
AJ500423 homologue to PIR|C84727|C8 probable glucan synthase [im... 136 9e-32
BQ147161 similar to PIR|T49914|T49 callose synthase catalytic su... 129 1e-29
AW696467 homologue to PIR|T49914|T4 callose synthase catalytic s... 100 2e-22
BG584489 similar to PIR|T47792|T4 hypothetical protein F17J16.15... 87 5e-17
TC83908 similar to PIR|T47792|T47792 hypothetical protein F17J16... 69 2e-11
TC84754 homologue to GP|7248390|dbj|BAA92713.1 ESTs AU033035(S15... 62 3e-09
BF644413 similar to GP|6642649|gb| putative glucan synthase {Ara... 52 2e-06
TC85053 similar to PIR|E85062|E85062 hypothetical protein AT4g04... 42 0.002
TC81545 similar to GP|15100055|gb|AAK84223.1 transcription facto... 33 0.84
TC86010 homologue to PIR|T00795|T00795 26S proteasome regulatory... 31 4.2
BG584727 weakly similar to PIR|T08399|T083 hypothetical protein ... 31 4.2
AJ498634 similar to GP|20196906|gb expressed protein {Arabidopsi... 31 4.2
BG586264 similar to PIR|T49193|T491 neoxanthin cleavage enzyme n... 30 9.3
TC83127 similar to PIR|T47697|T47697 Regulator of chromosome con... 30 9.3
TC76354 similar to GP|4589396|dbj|BAA76744.1 asparaginyl endopep... 30 9.3
>TC82856 homologue to PIR|T49914|T49914 callose synthase catalytic
subunit-like protein - Arabidopsis thaliana, partial
(12%)
Length = 764
Score = 496 bits (1276), Expect = e-140
Identities = 253/254 (99%), Positives = 253/254 (99%)
Frame = +3
Query: 1431 QLVRDCWPIP*GFVSTMGILMSLIGFFT*QEGVSVKPPRLSI*VKIFLLASTLHSVKEVL 1490
QLVRDCWPIP*GFVSTMGILMSLIGFFT*QEGVSVKPPRLSI*VKIFLLASTLHSVKEVL
Sbjct: 3 QLVRDCWPIP*GFVSTMGILMSLIGFFT*QEGVSVKPPRLSI*VKIFLLASTLHSVKEVL 182
Query: 1491 LIMSTYKLGREEMLVSTKFRCLKQK*LMEMESRH*VETCTGLGTVLISSECCPVILLQLD 1550
LIMSTYKLGREEMLVSTK RCLKQK*LMEMESRH*VETCTGLGTVLISSECCPVILLQLD
Sbjct: 183 LIMSTYKLGREEMLVSTKVRCLKQK*LMEMESRH*VETCTGLGTVLISSECCPVILLQLD 362
Query: 1551 FISALL*LFSLYTYFSMVASILFSVALKKG*VHKKPFETISLFKWLLLLSHSFKLGF*WP 1610
FISALL*LFSLYTYFSMVASILFSVALKKG*VHKKPFETISLFKWLLLLSHSFKLGF*WP
Sbjct: 363 FISALL*LFSLYTYFSMVASILFSVALKKG*VHKKPFETISLFKWLLLLSHSFKLGF*WP 542
Query: 1611 CPC*WKLAWKGASELHLVNLY*CSCSLLLYSSRFP*GPRLTILEGHCFMEVQNIDPRVED 1670
CPC*WKLAWKGASELHLVNLY*CSCSLLLYSSRFP*GPRLTILEGHCFMEVQNIDPRVED
Sbjct: 543 CPC*WKLAWKGASELHLVNLY*CSCSLLLYSSRFP*GPRLTILEGHCFMEVQNIDPRVED 722
Query: 1671 LWFSMPSLLIIIDF 1684
LWFSMPSLLIIIDF
Sbjct: 723 LWFSMPSLLIIIDF 764
>TC83029 similar to GP|20330757|gb|AAM19120.1 Putative glucan synthase {Oryza
sativa (japonica cultivar-group)}, partial (11%)
Length = 692
Score = 382 bits (980), Expect = e-105
Identities = 193/193 (100%), Positives = 193/193 (100%)
Frame = +3
Query: 1790 ES*WRYCCHCDFLSTNMVLYIT*ILQRRAQKVFWCMAFRGW*SL*YYL**RLYLLGGVNS 1849
ES*WRYCCHCDFLSTNMVLYIT*ILQRRAQKVFWCMAFRGW*SL*YYL**RLYLLGGVNS
Sbjct: 9 ES*WRYCCHCDFLSTNMVLYIT*ILQRRAQKVFWCMAFRGW*SL*YYL**RLYLLGGVNS 188
Query: 1850 VQIFSLSSD*SRG*YS*LLLRFWSF*SPFHI*HRKTLLFAFLPSCQLVGECYRLRKH*SL 1909
VQIFSLSSD*SRG*YS*LLLRFWSF*SPFHI*HRKTLLFAFLPSCQLVGECYRLRKH*SL
Sbjct: 189 VQIFSLSSD*SRG*YS*LLLRFWSF*SPFHI*HRKTLLFAFLPSCQLVGECYRLRKH*SL 368
Query: 1910 *YVVLDFGDL*KLLLVVMR*SWVCFCSLLLPFWLGFLLFQNFKPECCSTKRSAEVCKFLV 1969
*YVVLDFGDL*KLLLVVMR*SWVCFCSLLLPFWLGFLLFQNFKPECCSTKRSAEVCKFLV
Sbjct: 369 *YVVLDFGDL*KLLLVVMR*SWVCFCSLLLPFWLGFLLFQNFKPECCSTKRSAEVCKFLV 548
Query: 1970 FLEDRGRSALLAA 1982
FLEDRGRSALLAA
Sbjct: 549 FLEDRGRSALLAA 587
>TC89423 similar to GP|20330757|gb|AAM19120.1 Putative glucan synthase {Oryza
sativa (japonica cultivar-group)}, partial (17%)
Length = 1020
Score = 185 bits (469), Expect = 2e-46
Identities = 139/287 (48%), Positives = 163/287 (56%)
Frame = +2
Query: 1688 VTLSRVLSSWYYLLYTKFLVIHTEVLLHIY*LLYQCGSWWEPGCLLHSCSTPLGLNGKKL 1747
V L + L+ ++L T +L +HTE LHI+*L GSW G LL S S L+G++
Sbjct: 5 VILLKELNW*FFLWSTIYLAMHTEEWLHIF*LP*PYGSWQGHGSLLLSFSIHRDLSGRRY 184
Query: 1748 LMIGLIGINGLVTEEV*VYRLKKVGNLGGKKNKIIYSIQESGES*WRYCCHCDFLSTNMV 1807
LMIG IGI+G T EV VY LKK GN GGKK+ I SI E R H DF NM
Sbjct: 185 LMIGQIGISG*ATAEVSVYLLKKAGNPGGKKSMSILSIPECVVLLLR*YWHSDFSYINMD 364
Query: 1808 LYIT*ILQRRAQKVFWCMAFRGW*SL*YYL**RLYLLGGVNSVQIFSLSSD*SRG*YS*L 1867
LYIT KVFW M F G *S *Y++**R+YLL G SVQ SL D* R +S*
Sbjct: 365 LYIT-FQSPDHIKVFWFMVFHG**SF*YWV**RVYLLAGAASVQTSSLYLD*LRVLFS*H 541
Query: 1868 LLRFWSF*SPFHI*HRKTLLFAFLPSCQLVGECYRLRKH*SL*YVVLDFGDL*KLLLVVM 1927
L +* I* +T AFL SCQ GEC RL K S + LDFG + L V M
Sbjct: 542 FLPPS*Y**QSLI*QLRT**SAFLQSCQQAGECCRLHKLVSHSSLKLDFGARFEHLPVDM 721
Query: 1928 R*SWVCFCSLLLPFWLGFLLFQNFKPECCSTKRSAEVCKFLVFLEDR 1974
+* W C+ S W GF LFQ+FK EC STK S +V KFL FL DR
Sbjct: 722 K**WACYSSHQ*LSWHGFHLFQSFKQECFSTKLSVKVYKFLEFLVDR 862
>BI310709 similar to GP|20330757|g Putative glucan synthase {Oryza sativa
(japonica cultivar-group)}, partial (11%)
Length = 591
Score = 178 bits (451), Expect = 2e-44
Identities = 116/198 (58%), Positives = 133/198 (66%), Gaps = 2/198 (1%)
Frame = +1
Query: 1751 GLIGINGLVTEEV*VYRLKKVGNLGGKKNKIIYSIQESGES*WRYCCHCDFLSTNMVLYI 1810
GLIGINGL +E V*V LK+VGN GG++NK I +IQE ES* RYC C F + VL+
Sbjct: 1 GLIGINGLASEAV*VCHLKRVGNPGGRRNKSI*NIQECAES*PRYCYLCVFSFISTVLFT 180
Query: 1811 T*ILQRRAQKVFWCMAFRGW*SL*YYL**RLYLLGGVNSVQIFSLSSD*SRG*YS*LLLR 1870
T* L R QKV W MA+ GW*S *Y L *R LLG N VQIF+LSS * R *Y *LL +
Sbjct: 181 T*TL-RNLQKVSWSMAYHGW*SF*Y*LS*RRCLLGEGNLVQIFNLSSG**RD*YL*LLSQ 357
Query: 1871 FWSF*SPFHI*HRKTLLFAFLPSCQLVGECYRLRKH*SL*YVVLDFGDL*KLLLVVMR*S 1930
F * P * +TLLFAFLPSCQLVGECYRL KH*SL*Y +DFG+ *KL VVM+ S
Sbjct: 358 F**P*LPLLT*RCRTLLFAFLPSCQLVGECYRLHKH*SL*YAEVDFGNQ*KL*HVVMKLS 537
Query: 1931 W--VCFCSLLLPFWLGFL 1946
W + F + W F+
Sbjct: 538 WGLLLFTPVAFLAWFPFV 591
Score = 31.2 bits (69), Expect = 4.2
Identities = 13/16 (81%), Positives = 13/16 (81%)
Frame = +2
Query: 1932 VCFCSLLLPFWLGFLL 1947
VC CSLLLP W GFLL
Sbjct: 542 VCSCSLLLPSWPGFLL 589
>TC92382 similar to PIR|E86189|E86189 hypothetical protein [imported] -
Arabidopsis thaliana, partial (7%)
Length = 739
Score = 173 bits (438), Expect = 7e-43
Identities = 77/113 (68%), Positives = 96/113 (84%)
Frame = +3
Query: 535 AVLGVILSWKARRSMSLYVKLRYILKVISAAAWVILLSVTYAYTWDNPPGFAETIKSWFG 594
A+L ++LSWKAR MSL+VKLRYI K IS AAWV++L VTYA++W NP GF +TIK+WFG
Sbjct: 285 ALLDIVLSWKARNVMSLHVKLRYIFKAISGAAWVVILPVTYAFSWKNPSGFGQTIKNWFG 464
Query: 595 SNSSAPSLFIVAVVVYLSPNMLAAIFFMFPFIRRYLERSNYRIVMLMMWWSQI 647
+ S +PS+FI+AV +YLSPN+L+AI F+FPFIRRYLERSNY V LMMWWSQ+
Sbjct: 465 NGSGSPSIFILAVFIYLSPNILSAILFLFPFIRRYLERSNYGPVKLMMWWSQV 623
>AJ500423 homologue to PIR|C84727|C8 probable glucan synthase [imported] -
Arabidopsis thaliana, partial (12%)
Length = 581
Score = 136 bits (342), Expect = 9e-32
Identities = 97/193 (50%), Positives = 116/193 (59%), Gaps = 2/193 (1%)
Frame = -3
Query: 1305 KRSTRFTTLA**RLCRNPVVLQSLS--RIWTRSYIK*SFLVQRF*VRVSPKIKITP*FSH 1362
+R TRF * +L P L L ++ R + +*SF ++*VR S KIK P FS
Sbjct: 579 ER*TRFIIQH*PKLLYQPNQLIHLKQFKV*IR*FTE*SFPGLQY*VRGSQKIKTMPSFSQ 400
Query: 1363 VEKVCKQ*I*TRITTWKKP*K*EICCKNFLRSMMV*GSQVFLD*ESIYLLEVFLPLHGLC 1422
K CKQ I* RIT WKK K* ICCKNFLR+ + Q++LD ESI+LL VF LHGLC
Sbjct: 399 EAKACKQLI*IRITIWKKLSK*GICCKNFLRNTVAPDIQLYLDSESIFLLAVFHRLHGLC 220
Query: 1423 QIKRQVS*QLVRDCWPIP*GFVSTMGILMSLIGFFT*QEGVSVKPPRLSI*VKIFLLAST 1482
I++ V *QL RD W P* S M M LIGF T*QE V + R S * +I+L S
Sbjct: 219 PIRKPVL*QLDRDYWLTP*R*GSIMATQMYLIGFST*QEAV*ARLLRSST*ARIYLQVSI 40
Query: 1483 LHSVKEVLLIMST 1495
L VKE+ LIMST
Sbjct: 39 LLYVKEMSLIMST 1
>BQ147161 similar to PIR|T49914|T49 callose synthase catalytic subunit-like
protein - Arabidopsis thaliana, partial (4%)
Length = 266
Score = 129 bits (324), Expect = 1e-29
Identities = 68/91 (74%), Positives = 70/91 (76%), Gaps = 4/91 (4%)
Frame = +3
Query: 1671 LWFSMPSLLIIIDFIREVTLSRVLSS----WYYLLYTKFLVIHTEVLLHIY*LLYQCGSW 1726
LWFSMPSL D R + S + LLYTKFL IHTEVLLHIY*LLYQCGSW
Sbjct: 3 LWFSMPSLP---DNYRLYSRSHFVKGIEPLGITLLYTKFLXIHTEVLLHIY*LLYQCGSW 173
Query: 1727 WEPGCLLHSCSTPLGLNGKKLLMIGLIGING 1757
WEPGCLLHSCSTPLGLNGKKLLMIGLIGING
Sbjct: 174 WEPGCLLHSCSTPLGLNGKKLLMIGLIGING 266
>AW696467 homologue to PIR|T49914|T4 callose synthase catalytic subunit-like
protein - Arabidopsis thaliana, partial (6%)
Length = 511
Score = 100 bits (250), Expect(2) = 2e-22
Identities = 60/104 (57%), Positives = 70/104 (66%)
Frame = +2
Query: 1338 K*SFLVQRF*VRVSPKIKITP*FSHVEKVCKQ*I*TRITTWKKP*K*EICCKNFLRSMMV 1397
+*SF ++*VR S KIK P FS K CKQ I* RIT WKK K* ICCKNFLR+ +
Sbjct: 179 E*SFPGLQY*VRGSQKIKTMPSFSQEAKACKQLI*IRITIWKKLSK*GICCKNFLRNTVA 358
Query: 1398 *GSQVFLD*ESIYLLEVFLPLHGLCQIKRQVS*QLVRDCWPIP* 1441
Q++LD ESI+LL VF LHGLC I++ V *QL RD W P*
Sbjct: 359 PDIQLYLDSESIFLLAVFHRLHGLCPIRKPVL*QLDRDYWLTP* 490
Score = 25.0 bits (53), Expect(2) = 2e-22
Identities = 17/39 (43%), Positives = 25/39 (63%), Gaps = 1/39 (2%)
Frame = +1
Query: 1285 TLHL-ELPILMR*RNLSKTVRKRSTRFTTLA**RLCRNP 1322
++HL EL ILMR ++L+K + +R TRF * +L P
Sbjct: 22 SIHLSELLILMRLKSLAKILPER*TRFIIQH*PKLLYQP 138
>BG584489 similar to PIR|T47792|T4 hypothetical protein F17J16.150 -
Arabidopsis thaliana, partial (12%)
Length = 823
Score = 87.4 bits (215), Expect = 5e-17
Identities = 68/155 (43%), Positives = 75/155 (47%)
Frame = +1
Query: 1675 MPSLLIIIDFIREVTLSRVLSSWYYLLYTKFLVIHTEVLLHIY*LLYQCGSWWEPGCLLH 1734
+ SLLII EVTL R Y + K + HTE L I QCG W G LLH
Sbjct: 10 LQSLLIITGCTPEVTL*RASRYLYC*SFMKSMANHTEAQLFISSSQSQCGFWPFLGYLLH 189
Query: 1735 SCSTPLGLNGKKLLMIGLIGINGLVTEEV*VYRLKKVGNLGGKKNKIIYSIQESGES*WR 1794
SCS L GKK MIG+IG G T V V+ L K GNLGG K + S R
Sbjct: 190 SCSILQVLIGKKRWMIGVIGSGGWETVVVLVFHLIKAGNLGGMKKMNT*NTPMSEGKYLR 369
Query: 1795 YCCHCDFLSTNMVLYIT*ILQRRAQKVFWCMAFRG 1829
H LSTNM L T*IL R K+FW F G
Sbjct: 370 LFLHAASLSTNMELSTT*IL-RVVAKIFWYSHFLG 471
>TC83908 similar to PIR|T47792|T47792 hypothetical protein F17J16.150 -
Arabidopsis thaliana, partial (12%)
Length = 841
Score = 68.9 bits (167), Expect = 2e-11
Identities = 98/293 (33%), Positives = 124/293 (41%), Gaps = 4/293 (1%)
Frame = +1
Query: 1151 RYFQMSGPTFFSE*NVLARRNLKEMNLKS*KKSFVYGHHIEGRL*QEL*GV*CIIEKLWS 1210
RY M+GP S * V + LK++ + FV GH E R E *C I K +S
Sbjct: 1 RYIPMNGPILMSA*KV---KILKKIG----RSMFVSGHLTEDRHSLEQSEE*CTIGKPYS 159
Query: 1211 FRLSLIWPKTKI*WKATKPWKTQMITRGGKGHYGHSAKR*LI*NSHMLYHANNMALTSDL 1270
F S + KA T RG + K LI*+S ML+ N M L
Sbjct: 160 FSTS*KMRAIVVFLKAPGHLITMKEIRGL-----NKRKLWLI*SSPMLFLVNCMVHKKSL 324
Query: 1271 VL---PGHTTY*GL*LDTLHLELPILMR*RNLSKTVRKRSTRFTTLA**RLCRNPVVLQS 1327
+ T +* + + + K RK T F L
Sbjct: 325 KILLTEAATIIFSI*WSRTQPFGLLT*TKQKIQKGERKSITPF--------------L*R 462
Query: 1328 LSRIWTRSYIK*SFLV-QRF*VRVSPKIKITP*FSHVEKVCKQ*I*TRITTWKKP*K*EI 1386
R R YI SFLV QR V+ + K KI P + EK+C+ * RI T KK K*E+
Sbjct: 463 EGRSMIRKYIASSFLVLQRKLVKGNLKTKIMPLYLLAEKLCRTST*IRIITMKKLSK*EM 642
Query: 1387 CCKNFLRSMMV*GSQVFLD*ESIYLLEVFLPLHGLCQIKRQVS*QLVRDCWPI 1439
KN + G+Q+ *ESIYL EVF PL GLCQ +R * L + W I
Sbjct: 643 YWKNSTH-IRGKGNQLSWV*ESIYLPEVFHPLLGLCQTRRPAL*PLANEFWQI 798
>TC84754 homologue to GP|7248390|dbj|BAA92713.1 ESTs AU033035(S1515)
D39871(S1515) correspond to a region of the predicted
gene.~Similar to, partial (11%)
Length = 653
Score = 61.6 bits (148), Expect = 3e-09
Identities = 45/99 (45%), Positives = 56/99 (56%)
Frame = +3
Query: 1443 FVSTMGILMSLIGFFT*QEGVSVKPPRLSI*VKIFLLASTLHSVKEVLLIMSTYKLGREE 1502
F TMG M L +FT* VK L I VKI++LASTL E+ M+T++L +E
Sbjct: 12 FACTMGSQMYLTEYFT*LVEGLVKLHVLLILVKIYMLASTLRCGLEMSHTMNTFRLEKEG 191
Query: 1503 MLVSTKFRCLKQK*LMEMESRH*VETCTGLGTVLISSEC 1541
MLV T+ LK K* + MES+ TGL LIS EC
Sbjct: 192 MLV*TRLPSLKGK*QVVMESKCLAGIFTGLDNCLISLEC 308
>BF644413 similar to GP|6642649|gb| putative glucan synthase {Arabidopsis
thaliana}, partial (4%)
Length = 325
Score = 52.0 bits (123), Expect = 2e-06
Identities = 25/59 (42%), Positives = 38/59 (64%)
Frame = +1
Query: 166 SMEVDREILETQDKVAEKTEILVPFNILPLDPDSANQAIMKFPEIQAAVYALRNTRGLP 224
S E+ E+ + + TE L+ +NI+P+D ++ AI+ FPE+QAAV AL+ RGLP
Sbjct: 103 SEEIPDELKRVMESDSASTEDLIAYNIIPIDATTSTNAIVFFPEVQAAVPALKYFRGLP 279
>TC85053 similar to PIR|E85062|E85062 hypothetical protein AT4g04970
[imported] - Arabidopsis thaliana, partial (14%)
Length = 769
Score = 42.4 bits (98), Expect = 0.002
Identities = 29/71 (40%), Positives = 39/71 (54%)
Frame = +2
Query: 1471 SI*VKIFLLASTLHSVKEVLLIMSTYKLGREEMLVSTKFRCLKQK*LMEMESRH*VETCT 1530
SI VKIFLL S +H + IM+ Y+ +EEML ++ CL+ + + M SR * E T
Sbjct: 20 SILVKIFLLVSIVHCEVAM*HIMNIYR*VKEEMLA*IRYPCLRPRSQVAMASRC*AEMST 199
Query: 1531 GLGTVLISSEC 1541
G SS C
Sbjct: 200 G*DID*TSSAC 232
Score = 30.8 bits (68), Expect = 5.5
Identities = 18/43 (41%), Positives = 25/43 (57%)
Frame = +3
Query: 1653 LEGHCFMEVQNIDPRVEDLWFSMPSLLIIIDFIREVTLSRVLS 1695
L G FM V + +P+V LW+S LL I ++ E LSR L+
Sbjct: 561 LVGLYFMVVLSTEPQVAVLWWSTRVLLRTIGYMLEAILSRPLN 689
>TC81545 similar to GP|15100055|gb|AAK84223.1 transcription factor bZIP61
{Arabidopsis thaliana}, partial (12%)
Length = 596
Score = 33.5 bits (75), Expect = 0.84
Identities = 16/47 (34%), Positives = 26/47 (55%), Gaps = 7/47 (14%)
Frame = +1
Query: 1100 HCLW-----TCLWHQKFATCFHFRF*H--HTTQKRFSSHCLIWIRQM 1139
H LW TCL+ + +ATCF F + HT + HC+++I ++
Sbjct: 19 HSLWLLHIHTCLYKKTYATCFFILFVYLTHTLIHTHTLHCIVYINKI 159
>TC86010 homologue to PIR|T00795|T00795 26S proteasome regulatory subunit
[imported] - Arabidopsis thaliana, partial (35%)
Length = 1865
Score = 31.2 bits (69), Expect = 4.2
Identities = 15/40 (37%), Positives = 24/40 (59%), Gaps = 5/40 (12%)
Frame = +2
Query: 1671 LWFSMPSLLIIIDFIREVTLSRV-----LSSWYYLLYTKF 1705
+WF ++ I + +R++ LSRV SS YYL+Y +F
Sbjct: 1238 VWFLFSNVQIFVKVLRKLALSRVAT*MNCSSHYYLIYCRF 1357
>BG584727 weakly similar to PIR|T08399|T083 hypothetical protein F18B3.60 -
Arabidopsis thaliana, partial (7%)
Length = 854
Score = 31.2 bits (69), Expect = 4.2
Identities = 18/56 (32%), Positives = 31/56 (55%), Gaps = 3/56 (5%)
Frame = -3
Query: 1126 KRFSSHCLIWIRQMKMVFRF---CFICKRYFQMSGPTFFSE*NVLARRNLKEMNLK 1178
K F + + IR+M+ F F CFI +Y + + T+ S N+LA +LK + ++
Sbjct: 693 KPFRNSINLRIRRMQETFCFISHCFISLQYIKPTASTYISTINILAITDLKTIQIR 526
>AJ498634 similar to GP|20196906|gb expressed protein {Arabidopsis thaliana},
partial (28%)
Length = 520
Score = 31.2 bits (69), Expect = 4.2
Identities = 22/90 (24%), Positives = 38/90 (41%), Gaps = 15/90 (16%)
Frame = +3
Query: 334 GEAANLRFMPECLCYIYHHMAFEL---------YGMLAGNVSPMTGENIKPAYGG----- 379
GE N ++ ECL Y +HH+A ++ Y ++ G S GE+ Y
Sbjct: 252 GEEMNFTYV-ECLLYTFHHLAHKVPNATNSLCGYKIVTGQPSDRLGEDFSEQYDDFTERL 428
Query: 380 -EDEAFLRKVVTPIYNVIAELKKAKGEGQS 408
E F R + + +AE K+ + ++
Sbjct: 429 KNVEEFTRATIKKLTQGMAENNKSMADAKT 518
>BG586264 similar to PIR|T49193|T491 neoxanthin cleavage enzyme nc1 -
Arabidopsis thaliana, partial (17%)
Length = 660
Score = 30.0 bits (66), Expect = 9.3
Identities = 15/44 (34%), Positives = 28/44 (63%), Gaps = 1/44 (2%)
Frame = -3
Query: 144 LTKAYQTANVLFEVLKAVNMTQSME-VDREILETQDKVAEKTEI 186
LTK YQT N E+++ V ++ + +D E+L+ + V+E +E+
Sbjct: 310 LTKHYQTVNSWKEMIRVVELSHGKKAMDGEVLDRRSFVSEGSEV 179
>TC83127 similar to PIR|T47697|T47697 Regulator of chromosome
condensation-like protein - Arabidopsis thaliana, partial
(46%)
Length = 1208
Score = 30.0 bits (66), Expect = 9.3
Identities = 15/50 (30%), Positives = 23/50 (46%), Gaps = 2/50 (4%)
Frame = +2
Query: 1721 YQCGSWWE--PGCLLHSCSTPLGLNGKKLLMIGLIGINGLVTEEV*VYRL 1768
Y C SWWE PG + LGL ++ G +G++ +YR+
Sbjct: 155 YLCSSWWEAYPGTVRCGTGLGLGLWRRRTARFGFSSQDGIIPSSYFMYRV 304
>TC76354 similar to GP|4589396|dbj|BAA76744.1 asparaginyl endopeptidase
(VmPE-1) {Vigna mungo}, partial (91%)
Length = 1660
Score = 30.0 bits (66), Expect = 9.3
Identities = 11/31 (35%), Positives = 20/31 (64%)
Frame = +3
Query: 548 SMSLYVKLRYILKVISAAAWVILLSVTYAYT 578
S+ LY+++ +L V W+++L +TY YT
Sbjct: 423 SLLLYLEISQLLLVAVERLWIVVLMITYLYT 515
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.348 0.154 0.546
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 72,739,436
Number of Sequences: 36976
Number of extensions: 1227068
Number of successful extensions: 12256
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 5020
Number of HSP's successfully gapped in prelim test: 577
Number of HSP's that attempted gapping in prelim test: 6747
Number of HSP's gapped (non-prelim): 6593
length of query: 1982
length of database: 9,014,727
effective HSP length: 111
effective length of query: 1871
effective length of database: 4,910,391
effective search space: 9187341561
effective search space used: 9187341561
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 66 (30.0 bits)
Medicago: description of AC122723.7


