

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122723.6 - phase: 0
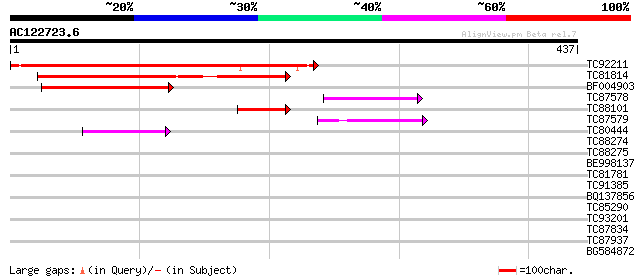
(437 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC92211 similar to PIR|B86182|B86182 hypothetical protein [impor... 290 6e-79
TC81814 weakly similar to GP|9294541|dbj|BAB02804.1 high mobilit... 168 4e-42
BF004903 similar to PIR|B96789|B96 protein T23E18.4 [imported] -... 130 1e-30
TC87578 similar to PIR|T12113|T12113 transcription factor - fava... 57 1e-08
TC88101 similar to PIR|B86182|B86182 hypothetical protein [impor... 50 1e-06
TC87579 homologue to PIR|T12113|T12113 transcription factor - fa... 45 6e-05
TC80444 similar to GP|15450515|gb|AAK96550.1 At2g17400 {Arabidop... 42 6e-04
TC88274 similar to GP|22022577|gb|AAM83245.1 AT5g54930/MBG8_20 {... 40 0.002
TC88275 similar to GP|22022577|gb|AAM83245.1 AT5g54930/MBG8_20 {... 38 0.007
BE998137 similar to GP|10177429|db zinc finger protein SHI-like ... 37 0.011
TC81781 similar to PIR|T49238|T49238 hypothetical protein F7K15.... 32 0.37
TC91385 similar to PIR|T49238|T49238 hypothetical protein F7K15.... 30 1.8
BQ137856 30 1.8
TC85290 similar to GP|7767653|gb|AAF69150.1| F27F5.2 {Arabidopsi... 29 3.1
TC93201 homologue to GP|17065164|gb|AAL32736.1 ADP-ribosylation ... 29 4.1
TC87834 similar to PIR|H84514|H84514 hypothetical protein At2g14... 28 5.3
TC87937 weakly similar to GP|4581119|gb|AAD24609.1| expressed pr... 28 7.0
BG584872 similar to PIR|T47766|T4 hypothetical protein F24I3.140... 28 7.0
>TC92211 similar to PIR|B86182|B86182 hypothetical protein [imported] -
Arabidopsis thaliana, partial (31%)
Length = 798
Score = 290 bits (743), Expect = 6e-79
Identities = 149/249 (59%), Positives = 188/249 (74%), Gaps = 11/249 (4%)
Frame = +3
Query: 1 MASTSCFFNNNEFPMKVVAPMSHSYPEPLAKYDDVVANPKLFMLTLEKLHASMGTKFMVP 60
MASTSCF + + PMK A YP P+A Y++VV NPKLF+L LEKLH MGTKFM+P
Sbjct: 57 MASTSCF-SKSPLPMKETALSHGEYPPPMATYEEVVDNPKLFILCLEKLHTLMGTKFMIP 233
Query: 61 IIGGKELDLCRLFIEVTSRGGIEKLMKERRWKEVTAAFSFPSTATNASFVLRKYYSSLLY 120
+IGG+ELDL RLF+EVTSRGG EK++K+R+WKEVT F+FPSTATNASFVLRKYY+SLLY
Sbjct: 234 VIGGRELDLHRLFVEVTSRGGFEKIIKDRKWKEVTLVFNFPSTATNASFVLRKYYTSLLY 413
Query: 121 HYEQIYYFRSKRWTPASSDALQNQSTMSVPASITQFLQPSPGTHPVDFQKSGVNAS--EL 178
HYEQIYYF+++ WT +SD LQ+QS++ PA QF PSP P FQ+ VN++ E
Sbjct: 414 HYEQIYYFKARDWTNTTSDVLQSQSSIPAPAPKMQFSHPSPQVQPAVFQQLKVNSAPPEA 593
Query: 179 PQVSSSGSSLAGVIDGKFESGYLVSVSVGSETLKGVLYESPQ---------SIKINNNNI 229
SS+GS + GVIDGKF+SGYLV+V++GSE LKGVLY++PQ S+ NNNN+
Sbjct: 594 MGSSSAGSQVVGVIDGKFDSGYLVTVTIGSEKLKGVLYQAPQNPVLPASHHSVPANNNNV 773
Query: 230 ASAALGVQR 238
+A++GV R
Sbjct: 774 -TASVGVLR 797
>TC81814 weakly similar to GP|9294541|dbj|BAB02804.1 high mobility group
protein-like {Arabidopsis thaliana}, partial (33%)
Length = 867
Score = 168 bits (425), Expect = 4e-42
Identities = 90/195 (46%), Positives = 119/195 (60%)
Frame = +1
Query: 22 SHSYPEPLAKYDDVVANPKLFMLTLEKLHASMGTKFMVPIIGGKELDLCRLFIEVTSRGG 81
++ YP P A Y D+V + LF L+ H S+GTK +P IGGK LDL LF+EVTSRGG
Sbjct: 172 TNPYPPPTAPYSDLVRDSNLFQQKLQSFHDSLGTKLKIPTIGGKPLDLHHLFVEVTSRGG 351
Query: 82 IEKLMKERRWKEVTAAFSFPSTATNASFVLRKYYSSLLYHYEQIYYFRSKRWTPASSDAL 141
IEK++ +R+WKEV +F+F T T+ SF++RK Y SLLYH+EQ YYF K+ P++ DAL
Sbjct: 352 IEKVIVDRKWKEVIMSFNFRDTITSGSFMVRKTYLSLLYHFEQAYYF-CKQVPPSTPDAL 528
Query: 142 QNQSTMSVPASITQFLQPSPGTHPVDFQKSGVNASELPQVSSSGSSLAGVIDGKFESGYL 201
S + T S V S + + GSS+ G ID KF+ GY+
Sbjct: 529 SGNVANSF----------TTNTDGAAINDSPVQVSPISPAQTLGSSVRGTIDMKFDDGYI 678
Query: 202 VSVSVGSETLKGVLY 216
V+V +GSE LKGVLY
Sbjct: 679 VTVDLGSEQLKGVLY 723
Score = 37.4 bits (85), Expect = 0.011
Identities = 17/30 (56%), Positives = 22/30 (72%)
Frame = +3
Query: 285 ISRMIGELWNNLKESEKTVYQEKAIKDKER 314
ISR IG + NL + E+ VYQEK ++DKER
Sbjct: 777 ISRKIGFMRRNLTDPERQVYQEKRVRDKER 866
>BF004903 similar to PIR|B96789|B96 protein T23E18.4 [imported] - Arabidopsis
thaliana, partial (30%)
Length = 709
Score = 130 bits (327), Expect = 1e-30
Identities = 60/102 (58%), Positives = 79/102 (76%)
Frame = +2
Query: 25 YPEPLAKYDDVVANPKLFMLTLEKLHASMGTKFMVPIIGGKELDLCRLFIEVTSRGGIEK 84
YP PLA ++DVV +P LF TL + H M TKFM+P+IGGKELDL L++EVT R G EK
Sbjct: 404 YPPPLASHNDVVNDPTLFWDTLRRFHFLMATKFMIPVIGGKELDLHVLYVEVTRRSGYEK 583
Query: 85 LMKERRWKEVTAAFSFPSTATNASFVLRKYYSSLLYHYEQIY 126
++ E++W+EV + F F ST T+ASFVLRK+Y +LLYHYEQ++
Sbjct: 584 VVAEKKWREVGSVFRFSSTTTSASFVLRKHYLNLLYHYEQVH 709
>TC87578 similar to PIR|T12113|T12113 transcription factor - fava bean,
partial (49%)
Length = 1021
Score = 57.0 bits (136), Expect = 1e-08
Identities = 27/77 (35%), Positives = 44/77 (57%), Gaps = 1/77 (1%)
Frame = +2
Query: 243 RKKSEIKRRDPAHPKPNRSGYNFFFAEQHARLKLLNQTMD-KDISRMIGELWNNLKESEK 301
+KK + K++DP PK SG+ FF + +K N + D+++++GE W + EK
Sbjct: 749 KKKKQKKKKDPNAPKRGMSGFMFFSQMERENIKKANPGISFTDVAKLLGENWKKMSAEEK 928
Query: 302 TVYQEKAIKDKERYQAE 318
Y+ KA DK+RY+ E
Sbjct: 929 EPYEAKARVDKKRYEDE 979
>TC88101 similar to PIR|B86182|B86182 hypothetical protein [imported] -
Arabidopsis thaliana, partial (7%)
Length = 1362
Score = 50.4 bits (119), Expect = 1e-06
Identities = 23/41 (56%), Positives = 30/41 (73%)
Frame = +3
Query: 176 SELPQVSSSGSSLAGVIDGKFESGYLVSVSVGSETLKGVLY 216
S + + GSS+ G IDGKF+ GY+V+V +GSE LKGVLY
Sbjct: 918 SPITPAQTLGSSVHGTIDGKFDDGYIVTVDLGSEQLKGVLY 1040
Score = 29.6 bits (65), Expect = 2.4
Identities = 12/21 (57%), Positives = 16/21 (76%)
Frame = +1
Query: 303 VYQEKAIKDKERYQAEMEDYR 323
VYQ+K ++DKERY EM Y+
Sbjct: 1081 VYQKKRLRDKERYGIEMLKYK 1143
>TC87579 homologue to PIR|T12113|T12113 transcription factor - fava bean,
partial (68%)
Length = 1778
Score = 45.1 bits (105), Expect = 6e-05
Identities = 28/85 (32%), Positives = 43/85 (49%)
Frame = +2
Query: 238 RRRRRRKKSEIKRRDPAHPKPNRSGYNFFFAEQHARLKLLNQTMDKDISRMIGELWNNLK 297
RR RRRK+ ++ R +F ++ KL+ + + R++GE W NL
Sbjct: 1043 RRNRRRKRIQMHLRGHC-----LVSCSFLRWKERI*RKLILEFH*RTWGRVLGEKWKNLS 1207
Query: 298 ESEKTVYQEKAIKDKERYQAEMEDY 322
EK Y+ KA +DK+RY+ EM Y
Sbjct: 1208 AEEKEPYEAKAQEDKKRYKDEMSGY 1282
>TC80444 similar to GP|15450515|gb|AAK96550.1 At2g17400 {Arabidopsis
thaliana}, partial (31%)
Length = 1118
Score = 41.6 bits (96), Expect = 6e-04
Identities = 22/68 (32%), Positives = 33/68 (48%)
Frame = +1
Query: 57 FMVPIIGGKELDLCRLFIEVTSRGGIEKLMKERRWKEVTAAFSFPSTATNASFVLRKYYS 116
F P G+ L+ +L+ V GG EK+ + W+ V +F P T T S+ R +Y
Sbjct: 34 FKHPKFYGEWLNCLKLWRAVMRLGGYEKVTSCKLWRSVGESFKPPKTCTTVSWTFRGFYE 213
Query: 117 SLLYHYEQ 124
L YE+
Sbjct: 214 KALLDYER 237
>TC88274 similar to GP|22022577|gb|AAM83245.1 AT5g54930/MBG8_20 {Arabidopsis
thaliana}, partial (30%)
Length = 1377
Score = 40.0 bits (92), Expect = 0.002
Identities = 29/87 (33%), Positives = 48/87 (54%), Gaps = 7/87 (8%)
Frame = +3
Query: 159 PSPGTHPVD----FQKSGVNASELPQVSSSGSSLAGVIDGKFESGYLVSVSVGSE--TLK 212
P PG V+ Q+ V S V G +++GVI+ F++GYL+SV VG+ TLK
Sbjct: 300 PPPGLQTVNGNQHLQRVQVRESNDVMV---GQAVSGVIEAVFDAGYLLSVRVGNSDTTLK 470
Query: 213 GVLYESPQSIKI-NNNNIASAALGVQR 238
G++++S + + N++A +QR
Sbjct: 471 GLVFKSGHFVPVCPENDVAPGIPMIQR 551
>TC88275 similar to GP|22022577|gb|AAM83245.1 AT5g54930/MBG8_20 {Arabidopsis
thaliana}, partial (20%)
Length = 599
Score = 38.1 bits (87), Expect = 0.007
Identities = 25/72 (34%), Positives = 41/72 (56%), Gaps = 6/72 (8%)
Frame = +3
Query: 159 PSPGTHPVD----FQKSGVNASELPQVSSSGSSLAGVIDGKFESGYLVSVSVGSE--TLK 212
P PG V+ Q+ V S V G +++GVI+ F++GYL+SV VG+ TLK
Sbjct: 345 PPPGLQTVNGNQHLQRVQVRESNDVMV---GQAVSGVIEAVFDAGYLLSVRVGNSDTTLK 515
Query: 213 GVLYESPQSIKI 224
G++++S + +
Sbjct: 516 GLVFKSGHFVPV 551
>BE998137 similar to GP|10177429|db zinc finger protein SHI-like {Arabidopsis
thaliana}, partial (36%)
Length = 592
Score = 37.4 bits (85), Expect = 0.011
Identities = 27/105 (25%), Positives = 47/105 (44%), Gaps = 9/105 (8%)
Frame = +1
Query: 133 WTPASSDALQNQSTMSVPASITQFLQPSPGTHPVDFQKS---GVNASELPQVSSSGSS-- 187
W PAS + Q S P +P G++ + ++ G+ + P V SS +
Sbjct: 142 WVPASRRRERQQQLSSSPLQRDISKRPRDGSNALVSTRNFHTGLEEANFPAVVSSPAEFR 321
Query: 188 ---LAGVIDGKFESGYLVSVSVGSETLKGVLYE-SPQSIKINNNN 228
++ + D Y +V++G KG+LY+ P+S N+NN
Sbjct: 322 CVRVSSIDDADDRYAYQTAVNIGGHLFKGILYDFGPESSTNNSNN 456
>TC81781 similar to PIR|T49238|T49238 hypothetical protein F7K15.90 -
Arabidopsis thaliana, partial (28%)
Length = 1027
Score = 32.3 bits (72), Expect = 0.37
Identities = 21/72 (29%), Positives = 33/72 (45%), Gaps = 4/72 (5%)
Frame = +2
Query: 56 KFMVPIIGGKELDLCRLFIEVTSRGGIEKLMKERRWK----EVTAAFSFPSTATNASFVL 111
+F I+ GK LDL L+ EV +RGG + WK ++ + T L
Sbjct: 449 EFPDAILNGKRLDLYNLYKEVVTRGGFH-VGNGINWKGQIFSKMGNYTSTNRMTGVGNTL 625
Query: 112 RKYYSSLLYHYE 123
+++Y + L YE
Sbjct: 626 KRHYETYLLEYE 661
>TC91385 similar to PIR|T49238|T49238 hypothetical protein F7K15.90 -
Arabidopsis thaliana, partial (29%)
Length = 726
Score = 30.0 bits (66), Expect = 1.8
Identities = 22/72 (30%), Positives = 33/72 (45%), Gaps = 4/72 (5%)
Frame = +2
Query: 56 KFMVPIIGGKELDLCRLFIEVTSRGGIEKLMKERRWK-EVTAAFS---FPSTATNASFVL 111
+F ++ K LDL L+ EV SRGG + WK +V + S T L
Sbjct: 230 EFPDAVLNAKRLDLFNLYREVVSRGGFH-VGNGINWKGQVFSKMSNHTLTHRMTGVGNTL 406
Query: 112 RKYYSSLLYHYE 123
+++Y + L YE
Sbjct: 407 KRHYETYLLEYE 442
>BQ137856
Length = 975
Score = 30.0 bits (66), Expect = 1.8
Identities = 16/46 (34%), Positives = 24/46 (51%), Gaps = 3/46 (6%)
Frame = +1
Query: 217 ESPQSIKINNNNIASAALGVQR---RRRRRKKSEIKRRDPAHPKPN 259
++PQ K + A G +R + R+ K + K RDP+HP PN
Sbjct: 130 QAPQPKKKTTPTLRREAFGTRRGMAKEPRQGK*QNKPRDPSHPSPN 267
>TC85290 similar to GP|7767653|gb|AAF69150.1| F27F5.2 {Arabidopsis
thaliana}, partial (32%)
Length = 1558
Score = 29.3 bits (64), Expect = 3.1
Identities = 18/60 (30%), Positives = 30/60 (50%), Gaps = 1/60 (1%)
Frame = +3
Query: 267 FAEQHARLKLLNQTMDKDI-SRMIGELWNNLKESEKTVYQEKAIKDKERYQAEMEDYRDK 325
F + + + N++ K+I I L KE E+ +EKA K+KER + E ++K
Sbjct: 639 FEDTQEYISIGNESYSKEIFEEYITYLKEKAKEKERKREEEKAKKEKEREEKEKRKEKEK 818
>TC93201 homologue to GP|17065164|gb|AAL32736.1 ADP-ribosylation factor-like
protein {Arabidopsis thaliana}, partial (95%)
Length = 683
Score = 28.9 bits (63), Expect = 4.1
Identities = 11/27 (40%), Positives = 17/27 (62%)
Frame = +2
Query: 108 SFVLRKYYSSLLYHYEQIYYFRSKRWT 134
SF+L K+ S +L+ YF +KRW+
Sbjct: 137 SFILNKWVSGILFSIGSAAYFLNKRWS 217
>TC87834 similar to PIR|H84514|H84514 hypothetical protein At2g14170
[imported] - Arabidopsis thaliana, partial (37%)
Length = 918
Score = 28.5 bits (62), Expect = 5.3
Identities = 14/40 (35%), Positives = 22/40 (55%)
Frame = -1
Query: 172 GVNASELPQVSSSGSSLAGVIDGKFESGYLVSVSVGSETL 211
G + LPQV+++G+ G+I GK + +GS TL
Sbjct: 744 GFRTNVLPQVTATGNIHNGIIAGKLKGQIPAQTPIGSLTL 625
>TC87937 weakly similar to GP|4581119|gb|AAD24609.1| expressed protein
{Arabidopsis thaliana}, partial (85%)
Length = 1172
Score = 28.1 bits (61), Expect = 7.0
Identities = 31/117 (26%), Positives = 51/117 (43%), Gaps = 3/117 (2%)
Frame = +2
Query: 289 IGELWNNLKESEK-TVYQEKAIKDKERY--QAEMEDYRDKMKTSIVTDNAGPLQQRFPEG 345
IG++ KESE TV++E + + A + D+ + I+ NA R+
Sbjct: 239 IGQISVPEKESEAGTVFEEATVLKETELAGNASLGMTIDRSRNRILVVNADVRGNRY--- 409
Query: 346 DSALVDVDIKMHDSCQTPEESSSGESDYVADDINMDASSGGARVDSEAFVDSDKATK 402
SALV D+ + S + +ADD+ +DA + A+V KA+K
Sbjct: 410 -SALVAYDLSTWKRLFLTQLSGPSDEKALADDVAVDA-------EGNAYVTDVKASK 556
>BG584872 similar to PIR|T47766|T4 hypothetical protein F24I3.140 -
Arabidopsis thaliana, partial (11%)
Length = 760
Score = 28.1 bits (61), Expect = 7.0
Identities = 11/27 (40%), Positives = 18/27 (65%)
Frame = +1
Query: 234 LGVQRRRRRRKKSEIKRRDPAHPKPNR 260
L +R R+RR +++R+P HP+P R
Sbjct: 145 LPTRRIRQRRLIRSLRQRNPLHPRPRR 225
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.313 0.129 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,975,481
Number of Sequences: 36976
Number of extensions: 149527
Number of successful extensions: 830
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 790
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 816
length of query: 437
length of database: 9,014,727
effective HSP length: 99
effective length of query: 338
effective length of database: 5,354,103
effective search space: 1809686814
effective search space used: 1809686814
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC122723.6


