

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122722.6 + phase: 0
(461 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
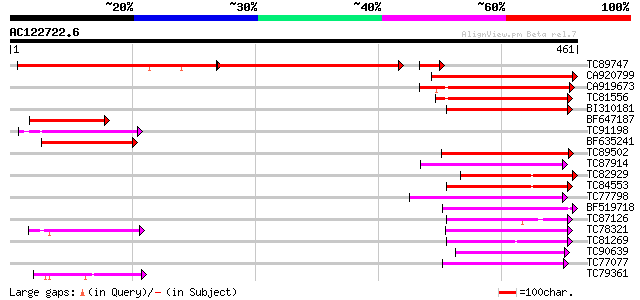
Score E
Sequences producing significant alignments: (bits) Value
TC89747 similar to PIR|F96696|F96696 protein F1N21.12 [imported]... 294 e-157
CA920799 similar to PIR|F96696|F96 protein F1N21.12 [imported] -... 230 8e-61
CA919673 similar to GP|9858106|gb| putative glucose translocator... 124 8e-29
TC81556 similar to GP|2688830|gb|AAB88879.1| putative sugar tran... 117 9e-27
BI310181 similar to PIR|C86184|C86 hypothetical protein [importe... 101 5e-22
BF647187 similar to PIR|F96696|F966 protein F1N21.12 [imported] ... 99 3e-21
TC91198 similar to GP|16648753|gb|AAL25568.1 AT5g16150/T21H19_70... 80 1e-15
BF635241 similar to GP|8347250|gb| hexose transporter {Arabidops... 77 2e-14
TC89502 similar to GP|9759246|dbj|BAB09770.1 sugar transporter-l... 74 1e-13
TC87914 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_14... 70 2e-12
TC82929 similar to PIR|D96589|D96589 hypothetical protein T22H22... 69 4e-12
TC84553 similar to GP|17381265|gb|AAL36051.1 AT5g18840/F17K4_90 ... 68 7e-12
TC77798 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_14... 68 8e-12
BF519718 weakly similar to PIR|G84564|G84 probable sugar transpo... 62 4e-10
TC87126 similar to GP|15724240|gb|AAL06513.1 At1g75220/F22H5_6 {... 62 4e-10
TC78321 similar to GP|21689841|gb|AAM67564.1 unknown protein {Ar... 61 1e-09
TC81269 similar to GP|23504385|emb|CAC00697. putative sugar tran... 60 1e-09
TC90639 similar to GP|17380890|gb|AAL36257.1 putative membrane t... 60 2e-09
TC77077 similar to GP|9293909|dbj|BAB01812.1 sugar transporter p... 60 2e-09
TC79361 similar to GP|17380890|gb|AAL36257.1 putative membrane t... 52 4e-07
>TC89747 similar to PIR|F96696|F96696 protein F1N21.12 [imported] -
Arabidopsis thaliana, partial (64%)
Length = 1372
Score = 294 bits (752), Expect(3) = e-157
Identities = 148/151 (98%), Positives = 150/151 (99%)
Frame = +1
Query: 170 ISGCLPYLQLYLLWLWSSVQRVHIGYTSKEELLKQKLSLKDFWVYPKQNLRCHSYPR*IE 229
+SG LPYLQLYLLWLW+SVQRVHIGYTSKEELLKQKLSLKDFWVYPKQNLRCHSYPR*IE
Sbjct: 856 VSGYLPYLQLYLLWLWTSVQRVHIGYTSKEELLKQKLSLKDFWVYPKQNLRCHSYPR*IE 1035
Query: 230 EKILTL*SSLNCFMAIILKLFLLDQPYLLYNSYLV*TLCFISLQLFLKVPECHRILQTSA 289
EKILTL*SSLNCFMAIILKLFLLDQPYLLYNSYLV*TLCFISLQLFLKVPECHRILQTSA
Sbjct: 1036 EKILTL*SSLNCFMAIILKLFLLDQPYLLYNSYLV*TLCFISLQLFLKVPECHRILQTSA 1215
Query: 290 *ELRI*QDLSFRRV*WINLEGRFYSFGVSLA 320
*ELRI*QDLSFRRV*WINLEGRFYSFGVSLA
Sbjct: 1216 *ELRI*QDLSFRRV*WINLEGRFYSFGVSLA 1308
Score = 261 bits (667), Expect(3) = e-157
Identities = 139/176 (78%), Positives = 143/176 (80%), Gaps = 10/176 (5%)
Frame = +3
Query: 7 IDLLDNFIDKETTNPSWKLSLPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLA 66
+DLLDNFIDKETTNPSWKLSLPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLA
Sbjct: 315 LDLLDNFIDKETTNPSWKLSLPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLA 494
Query: 67 EGLVVSICLGGALFGCLLSGWIADAVGRRRAFQLCALPMIIGAAMS--NKQLVWHACRKI 124
EGLVVSICLGGALFGCLLSGWIADAVGRRRAFQLCALPMIIGAAMS L ++
Sbjct: 495 EGLVVSICLGGALFGCLLSGWIADAVGRRRAFQLCALPMIIGAAMSAATNNLFGMLVGRL 674
Query: 125 ICWNWLGLGPSRCC--------SLCDRAYGALIQIATCFGILGSLFIGIPVKEISG 172
LGLGP + YGALIQIATCFGILGSLFIGIPVKEISG
Sbjct: 675 FVGTGLGLGPPVAALYVTEVSPAFVRGTYGALIQIATCFGILGSLFIGIPVKEISG 842
Score = 40.0 bits (92), Expect(3) = e-157
Identities = 20/20 (100%), Positives = 20/20 (100%)
Frame = +3
Query: 334 AISMIIQATGASTLLPTAGA 353
AISMIIQATGASTLLPTAGA
Sbjct: 1311 AISMIIQATGASTLLPTAGA 1370
>CA920799 similar to PIR|F96696|F96 protein F1N21.12 [imported] - Arabidopsis
thaliana, partial (22%)
Length = 752
Score = 230 bits (587), Expect = 8e-61
Identities = 118/118 (100%), Positives = 118/118 (100%)
Frame = -1
Query: 344 ASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVI 403
ASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVI
Sbjct: 752 ASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVI 573
Query: 404 NFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIEIALLPQE 461
NFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIEIALLPQE
Sbjct: 572 NFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIEIALLPQE 399
>CA919673 similar to GP|9858106|gb| putative glucose translocator
{Mesembryanthemum crystallinum}, partial (31%)
Length = 620
Score = 124 bits (311), Expect = 8e-29
Identities = 64/128 (50%), Positives = 91/128 (71%), Gaps = 2/128 (1%)
Frame = -3
Query: 334 AISMIIQATGAS--TLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAK 391
A SM++ + S L P +G+L +V G +L+V +F+LGAGPVP LLL EIF SRIRAK
Sbjct: 477 AASMLLLSVSFSWKVLAPYSGSL--AVLGTVLYVLSFSLGAGPVPALLLPEIFASRIRAK 304
Query: 392 AMAFCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQ 451
A++ + HW+ NF +GL FL ++ K+G +Y F+T C++AV+++ NVVETKG+SL+
Sbjct: 303 AISLSLGTHWISNFVIGLYFLSVVNKIGISSVYLGFSTVCLLAVLYIAANVVETKGRSLE 124
Query: 452 EIEIALLP 459
EIE AL P
Sbjct: 123 EIERALTP 100
>TC81556 similar to GP|2688830|gb|AAB88879.1| putative sugar transporter
{Prunus armeniaca}, partial (59%)
Length = 1074
Score = 117 bits (293), Expect = 9e-27
Identities = 58/111 (52%), Positives = 80/111 (71%)
Frame = +1
Query: 347 LLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFF 406
L P +G L +V G +L+V +F+LGAGPVP LLL EIF SRIRAKA++ + HW+ NF
Sbjct: 532 LAPYSGTL--AVLGTVLYVLSFSLGAGPVPALLLPEIFASRIRAKAVSLSLGTHWISNFV 705
Query: 407 VGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIEIAL 457
+GL FL ++ K G +Y F+ C++AV+++ NVVETKG+SL+EIE AL
Sbjct: 706 IGLYFLSVVNKFGISSVYLGFSAVCVLAVLYIAGNVVETKGRSLEEIERAL 858
>BI310181 similar to PIR|C86184|C86 hypothetical protein [imported] -
Arabidopsis thaliana, partial (18%)
Length = 656
Score = 101 bits (252), Expect = 5e-22
Identities = 48/102 (47%), Positives = 68/102 (66%)
Frame = +2
Query: 356 LSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLLFLRLL 415
LS+ G ++++F+FA+GAGPV G+++ E+ +R R K M F S HWV NF VGL FL L+
Sbjct: 53 LSILGTIMYIFSFAIGAGPVTGIIIPELSSTRTRGKIMGFSFSTHWVCNFVVGLFFLELV 232
Query: 416 EKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIEIAL 457
EK G +Y+ F ++A F +VETKG SL+EIE +L
Sbjct: 233 EKFGVAPVYASFGAVSLLAAAFAHYFLVETKGCSLEEIERSL 358
>BF647187 similar to PIR|F96696|F966 protein F1N21.12 [imported] -
Arabidopsis thaliana, partial (12%)
Length = 499
Score = 99.4 bits (246), Expect = 3e-21
Identities = 45/65 (69%), Positives = 56/65 (85%)
Frame = +1
Query: 17 ETTNPSWKLSLPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICLG 76
++ P W+ SL HV+VA+I+SFL+GYH+GVVNE LESIS+DLGF+G+TLAEGLV SICLG
Sbjct: 301 QSAKPCWRRSLRHVIVASISSFLYGYHIGVVNETLESISIDLGFSGDTLAEGLVXSICLG 480
Query: 77 GALFG 81
GA G
Sbjct: 481 GAFIG 495
>TC91198 similar to GP|16648753|gb|AAL25568.1 AT5g16150/T21H19_70
{Arabidopsis thaliana}, partial (31%)
Length = 673
Score = 80.5 bits (197), Expect = 1e-15
Identities = 48/101 (47%), Positives = 60/101 (58%)
Frame = +2
Query: 8 DLLDNFIDKETTNPSWKLSLPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAE 67
DLL N K PS + P+V VA + +FLFGYHLGVVN LE ++ DL NT+ +
Sbjct: 179 DLLPN---KSPGRPSGTV-FPYVGVACLGAFLFGYHLGVVNGALEYLAKDLRIAQNTVLQ 346
Query: 68 GLVVSICLGGALFGCLLSGWIADAVGRRRAFQLCALPMIIG 108
G +VS L GA G G +AD GR R FQL A+P+ IG
Sbjct: 347 GWIVSTLLAGATVGSFTGGALADKFGRTRTFQLDAIPLAIG 469
>BF635241 similar to GP|8347250|gb| hexose transporter {Arabidopsis
thaliana}, partial (21%)
Length = 672
Score = 76.6 bits (187), Expect = 2e-14
Identities = 40/78 (51%), Positives = 50/78 (63%)
Frame = +1
Query: 27 LPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICLGGALFGCLLSG 86
LP+V VA + + LFGYHLGVVN LE ++ DLG NT+ +G +VS L GA G G
Sbjct: 439 LPYVGVACLGAILFGYHLGVVNGALEYLAKDLGIVENTVLQGWIVSSLLAGATVGSFTGG 618
Query: 87 WIADAVGRRRAFQLCALP 104
+AD GR R FQL A+P
Sbjct: 619 TLADKFGRTRTFQLDAIP 672
>TC89502 similar to GP|9759246|dbj|BAB09770.1 sugar transporter-like protein
{Arabidopsis thaliana}, partial (27%)
Length = 595
Score = 73.9 bits (180), Expect = 1e-13
Identities = 37/107 (34%), Positives = 65/107 (60%)
Frame = +2
Query: 352 GALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLLF 411
G ++V +LL+V + + GP+ L+++EIFP R R + ++ + ++ N V F
Sbjct: 131 GLPIVAVAALLLYVGCYQISFGPISWLMVSEIFPLRTRGRGISMAVLTNFAANAVVTFAF 310
Query: 412 LRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIEIALL 458
L E LGA+ L+ +FA +++++F+ +V ETKG SL+EIE +L
Sbjct: 311 SPLKEYLGAENLFLLFAAIALVSLVFIVTSVPETKGLSLEEIESKIL 451
>TC87914 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_140
{Arabidopsis thaliana}, partial (39%)
Length = 1129
Score = 70.1 bits (170), Expect = 2e-12
Identities = 36/119 (30%), Positives = 61/119 (51%)
Frame = +3
Query: 335 ISMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMA 394
+S+ I G+ L +S ++++ +F +G GPVP +L EIFP+R+R +A
Sbjct: 546 VSLFILVLGSLVDLGDTANASISTISVVVYFCSFVMGFGPVPNILCAEIFPTRVRGLCIA 725
Query: 395 FCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEI 453
C W+ + V +L +G ++ ++A C +A +FV V ETKG L+ I
Sbjct: 726 ICALTFWICDIIVTYSLPVMLNSVGLGGVFGLYAVVCCIAWVFVFLKVPETKGMPLEVI 902
>TC82929 similar to PIR|D96589|D96589 hypothetical protein T22H22.15
[imported] - Arabidopsis thaliana, partial (30%)
Length = 779
Score = 68.9 bits (167), Expect = 4e-12
Identities = 32/95 (33%), Positives = 60/95 (62%)
Frame = +1
Query: 367 TFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLLFLRLLEKLGAQLLYSM 426
+F+LG G +P ++++EIFP ++ A +F VHW+ ++ V F L+ A + +
Sbjct: 466 SFSLGMGGIPWVIMSEIFPINVKGSAGSFVTFVHWLCSWIVSYAFNFLMSWNSAGTFF-I 642
Query: 427 FATFCIMAVIFVKRNVVETKGKSLQEIEIALLPQE 461
F+T C + ++FV + V ETKG++L+E++ +L P +
Sbjct: 643 FSTICGLTILFVAKLVPETKGRTLEEVQASLNPYQ 747
>TC84553 similar to GP|17381265|gb|AAL36051.1 AT5g18840/F17K4_90
{Arabidopsis thaliana}, partial (41%)
Length = 827
Score = 68.2 bits (165), Expect = 7e-12
Identities = 34/102 (33%), Positives = 63/102 (61%)
Frame = +2
Query: 356 LSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLLFLRLL 415
L+V G+L++V F++G GPVP ++++EIFP ++ A + + ++W+ + V F L
Sbjct: 302 LAVAGILIYVAAFSIGMGPVPWVIMSEIFPIHVKGTAGSLVVLINWLGAWVVSYTFNFFL 481
Query: 416 EKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIEIAL 457
L+ ++A ++ ++FV + V ETKGK+L+EI+ L
Sbjct: 482 SWSSPGTLF-LYAGCSLLTILFVAKLVPETKGKTLEEIQACL 604
>TC77798 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_140
{Arabidopsis thaliana}, partial (88%)
Length = 2467
Score = 67.8 bits (164), Expect = 8e-12
Identities = 37/128 (28%), Positives = 66/128 (50%)
Frame = +3
Query: 326 LYIFCTV*AISMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFP 385
L + V +S++I G+ + +S ++++ F +G GP+P +L +EIFP
Sbjct: 1866 LLVTIPVLIVSLVILVLGSVIDFGSVVHAAISTVCVVVYFCFFVMGYGPIPNILCSEIFP 2045
Query: 386 SRIRAKAMAFCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVET 445
+R+R +A C V W+ + V +L LG ++ ++A C ++ +FV V ET
Sbjct: 2046 TRVRGLCIAICALVFWIGDIIVTYSLPVMLSSLGLAGVFGVYAIVCCISWVFVYLKVPET 2225
Query: 446 KGKSLQEI 453
KG L+ I
Sbjct: 2226 KGMPLEVI 2249
Score = 35.0 bits (79), Expect = 0.061
Identities = 27/96 (28%), Positives = 47/96 (48%)
Frame = +3
Query: 31 LVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICLGGALFGCLLSGWIAD 90
+ A+I +FL G+ + + I DL T EGLVV++ L GA SG I+D
Sbjct: 135 IAASIGNFLQGWDNATIAGSILYIKKDLALQ--TTMEGLVVAMSLIGATVITTCSGPISD 308
Query: 91 AVGRRRAFQLCALPMIIGAAMSNKQLVWHACRKIIC 126
+GRR + ++ +G+ + ++W ++C
Sbjct: 309 WLGRRPMMIISSVLYFLGSLV----MLWSPNVYVLC 404
>BF519718 weakly similar to PIR|G84564|G84 probable sugar transporter
[imported] - Arabidopsis thaliana, partial (20%)
Length = 659
Score = 62.4 bits (150), Expect = 4e-10
Identities = 38/109 (34%), Positives = 64/109 (57%)
Frame = +3
Query: 353 ALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLLFL 412
A+ L V + V F++G GP + +EIFP R+RA+ + +SV+ +I+ V + FL
Sbjct: 174 AIALCVVAVCAAVSFFSIGLGPTTWVYSSEIFPMRLRAQGTSLAISVNRLISGVVSMSFL 353
Query: 413 RLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIEIALLPQE 461
+ E++ ++ + A ++A +F + ETKGKSL+EIE AL +E
Sbjct: 354 SISEEITFGGMFFVLAGVMVLATLFFYYFLPETKGKSLEEIE-ALFEEE 497
>TC87126 similar to GP|15724240|gb|AAL06513.1 At1g75220/F22H5_6 {Arabidopsis
thaliana}, partial (79%)
Length = 1524
Score = 62.4 bits (150), Expect = 4e-10
Identities = 35/104 (33%), Positives = 61/104 (58%), Gaps = 2/104 (1%)
Frame = +3
Query: 356 LSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLLFLRLL 415
+SV G+++ V F+LG GP+P L+++EI P I+ A + +W++ + + + LL
Sbjct: 873 ISVVGLVVMVIGFSLGLGPIPWLIMSEILPVNIKGLAGSTATMANWLVAWIITMTANLLL 1052
Query: 416 --EKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIEIAL 457
G L+Y++ A F V+F V ETKG++L+EI+ +L
Sbjct: 1053TWSSGGTFLIYTVVAAF---TVVFTSLWVPETKGRTLEEIQFSL 1175
Score = 33.1 bits (74), Expect = 0.23
Identities = 14/32 (43%), Positives = 22/32 (68%)
Frame = +3
Query: 77 GALFGCLLSGWIADAVGRRRAFQLCALPMIIG 108
GA+ G + SG IA+ VGR+ + + ++P IIG
Sbjct: 6 GAMVGAIASGQIAEYVGRKGSLMIASIPNIIG 101
>TC78321 similar to GP|21689841|gb|AAM67564.1 unknown protein {Arabidopsis
thaliana}, partial (80%)
Length = 2093
Score = 60.8 bits (146), Expect = 1e-09
Identities = 33/103 (32%), Positives = 54/103 (52%)
Frame = +2
Query: 355 YLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLLFLRL 414
++++ + L++ F+ G G VP ++ +EI+P R R + WV N V FL L
Sbjct: 1487 WIAILALALYIIFFSPGMGTVPWVVNSEIYPLRYRGICGGIASTTVWVSNLVVSQSFLSL 1666
Query: 415 LEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIEIAL 457
+G + +FA I+A+ FV V ETKG ++E+E L
Sbjct: 1667 TVAIGPAWTFMIFAIIAIVAIFFVIIFVPETKGVPMEEVESML 1795
Score = 42.4 bits (98), Expect = 4e-04
Identities = 30/99 (30%), Positives = 47/99 (47%), Gaps = 5/99 (5%)
Frame = +2
Query: 16 KETTNPSWKLSLPHVL----VATITSFLFGYHLGVVNEPLESISVDL-GFNGNTLAEGLV 70
+E + SWK P+VL A I LFGY GV++ L I + T + +
Sbjct: 185 RECLSLSWKN--PYVLRLAFSAGIGGLLFGYDTGVISGALLYIRDEFPAVEKKTWLQEAI 358
Query: 71 VSICLGGALFGCLLSGWIADAVGRRRAFQLCALPMIIGA 109
VS + GA+ G + GWI D GR+ + + ++G+
Sbjct: 359 VSTAIAGAIIGAAIGGWINDRFGRKVSIIVADTLFLLGS 475
>TC81269 similar to GP|23504385|emb|CAC00697. putative sugar transporter
{Lycopersicon esculentum}, partial (42%)
Length = 966
Score = 60.5 bits (145), Expect = 1e-09
Identities = 30/102 (29%), Positives = 61/102 (59%)
Frame = +2
Query: 356 LSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLLFLRLL 415
L+V G+L+++ +F++G G +P ++++EIFP I+ +A + V+W + F L
Sbjct: 332 LAVTGILVYIGSFSIGMGAIPWVVMSEIFPVNIKGQAGSIATLVNWFGAWLCSYTF-NFL 508
Query: 416 EKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIEIAL 457
+ + ++A +A++F+ V ETKGKSL++++ A+
Sbjct: 509 MSWSSYGTFVLYAAINALAILFIAVVVPETKGKSLEQLQAAI 634
>TC90639 similar to GP|17380890|gb|AAL36257.1 putative membrane transporter
protein {Arabidopsis thaliana}, partial (21%)
Length = 555
Score = 60.1 bits (144), Expect = 2e-09
Identities = 29/93 (31%), Positives = 49/93 (52%)
Frame = +2
Query: 363 LFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLLFLRLLEKLGAQL 422
L++ F+ G GPVPG + +EI+P R +V W+ N V FL + + +G
Sbjct: 5 LYIGFFSPGMGPVPGTINSEIYPEEYRGICGGMAATVCWISNLIVSESFLSIADAIGIAS 184
Query: 423 LYSMFATFCIMAVIFVKRNVVETKGKSLQEIEI 455
+ + A ++A +FV V ET+G + E+E+
Sbjct: 185 TFLIIAVIAVVAFLFVLLYVPETQGLTFDEVEL 283
>TC77077 similar to GP|9293909|dbj|BAB01812.1 sugar transporter protein
{Arabidopsis thaliana}, partial (85%)
Length = 1752
Score = 59.7 bits (143), Expect = 2e-09
Identities = 34/102 (33%), Positives = 60/102 (58%)
Frame = +1
Query: 353 ALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLLFL 412
A+ LS+ +L +V TF++GAGP+ + +EIFP R+RA+ A + V+ V + + + FL
Sbjct: 1180 AIGLSIATVLSYVATFSIGAGPITWVYSSEIFPLRLRAQGAACGVVVNRVTSGVISMTFL 1359
Query: 413 RLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIE 454
L + + + +F + IF + ET+GK+L+E+E
Sbjct: 1360 SLSKGITIGGAFFLFGGIATIGWIFFYIMLPETQGKTLEEME 1485
Score = 33.9 bits (76), Expect = 0.14
Identities = 23/88 (26%), Positives = 42/88 (47%)
Frame = +1
Query: 24 KLSLPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICLGGALFGCL 83
K + ++A++TS L GY +GV++ I DL + + +++ I +L G
Sbjct: 103 KYAFACAMLASMTSILLGYDIGVMSGAAIYIKRDLKVSDGKIE--VLLGIINIYSLIGSC 276
Query: 84 LSGWIADAVGRRRAFQLCALPMIIGAAM 111
L+G +D +GRR +GA +
Sbjct: 277 LAGRTSDWIGRRYTIVFAGAIFFVGALL 360
>TC79361 similar to GP|17380890|gb|AAL36257.1 putative membrane transporter
protein {Arabidopsis thaliana}, partial (55%)
Length = 949
Score = 52.4 bits (124), Expect = 4e-07
Identities = 37/101 (36%), Positives = 53/101 (51%), Gaps = 9/101 (8%)
Frame = +1
Query: 20 NPSWKLSL---PHVL----VATITSFLFGYHLGVVNEPLESISVDLGF--NGNTLAEGLV 70
+P K+S P++L VA I LFGY GV++ L I D N N L E +
Sbjct: 130 HPDRKMSFFKNPYILGLAAVAGIGGLLFGYDTGVISGALLYIKDDFPQVRNSNFLQE-TI 306
Query: 71 VSICLGGALFGCLLSGWIADAVGRRRAFQLCALPMIIGAAM 111
VS+ + GA+ G GW+ DA GR++A L + I+GA +
Sbjct: 307 VSMAIAGAIVGAAFGGWLNDAYGRKKATLLADVIFILGAIL 429
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.341 0.151 0.500
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,620,923
Number of Sequences: 36976
Number of extensions: 298560
Number of successful extensions: 2571
Number of sequences better than 10.0: 84
Number of HSP's better than 10.0 without gapping: 2528
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2559
length of query: 461
length of database: 9,014,727
effective HSP length: 99
effective length of query: 362
effective length of database: 5,354,103
effective search space: 1938185286
effective search space used: 1938185286
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC122722.6


