

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122722.5 - phase: 0
(379 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
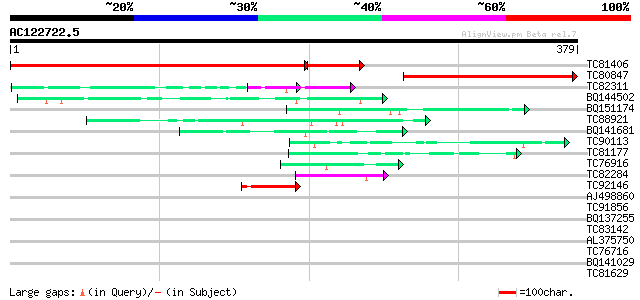
Score E
Sequences producing significant alignments: (bits) Value
TC81406 weakly similar to PIR|T08954|T08954 hypothetical protein... 446 e-146
TC80847 similar to GP|23326831|gb|AAN25338.1 polyribonucleotide ... 253 7e-68
TC82311 weakly similar to PIR|G96696|G96696 protein F1N21.14 [im... 47 1e-05
BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 45 4e-05
BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xy... 44 1e-04
TC88921 similar to SP|P92966|RS41_ARATH Arginine/serine-rich spl... 43 2e-04
BQ141681 weakly similar to GP|15145793|gb| basic proline-rich pr... 42 3e-04
TC90113 similar to PIR|C86301|C86301 arginine/serine-rich protei... 42 3e-04
TC81177 similar to GP|15293081|gb|AAK93651.1 unknown protein {Ar... 42 4e-04
TC76916 MtN4 41 7e-04
TC82284 weakly similar to GP|11119510|gb|AAF37724.2 diaphanous p... 41 9e-04
TC92146 weakly similar to PIR|T37870|T37870 RNA-binding / Ran zi... 41 9e-04
AJ498860 similar to GP|20197423|gb|A unknown protein {Arabidopsi... 40 0.001
TC91856 similar to GP|11994340|dbj|BAB02299. zinc finger protein... 39 0.003
BQ137255 homologue to PIR|B34768|B347 ORF5 protein - Orf virus (... 39 0.003
TC83142 similar to GP|20197597|gb|AAM15145.1 predicted protein {... 39 0.003
AL375750 weakly similar to PIR|A96592|A965 hypothetical protein ... 38 0.007
TC76716 similar to PIR|T10863|T10863 extensin precursor - kidney... 38 0.007
BQ141029 similar to GP|8132441|gb| extensin {Pisum sativum}, par... 37 0.010
TC81629 similar to GP|15081654|gb|AAK82482.1 AT4g26750/F10M23_90... 37 0.010
>TC81406 weakly similar to PIR|T08954|T08954 hypothetical protein F19B15.20
- Arabidopsis thaliana, partial (20%)
Length = 747
Score = 446 bits (1146), Expect(2) = e-146
Identities = 200/200 (100%), Positives = 200/200 (100%)
Frame = +3
Query: 1 MSSTSPSQSPPSHHHHHQPLLSSLVVRPSLTENSPPGAAAHSNDYEPGELRRDPPPVNYS 60
MSSTSPSQSPPSHHHHHQPLLSSLVVRPSLTENSPPGAAAHSNDYEPGELRRDPPPVNYS
Sbjct: 36 MSSTSPSQSPPSHHHHHQPLLSSLVVRPSLTENSPPGAAAHSNDYEPGELRRDPPPVNYS 215
Query: 61 RSDRYSDDAGYRFRAGSSSPVHHRRDADHRFPSDYNHFPRNRGFGGGRDFGRFRDPPHPY 120
RSDRYSDDAGYRFRAGSSSPVHHRRDADHRFPSDYNHFPRNRGFGGGRDFGRFRDPPHPY
Sbjct: 216 RSDRYSDDAGYRFRAGSSSPVHHRRDADHRFPSDYNHFPRNRGFGGGRDFGRFRDPPHPY 395
Query: 121 ARGRPGGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNVRPREGDWMCPDALCGNLNFARRD 180
ARGRPGGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNVRPREGDWMCPDALCGNLNFARRD
Sbjct: 396 ARGRPGGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNVRPREGDWMCPDALCGNLNFARRD 575
Query: 181 FCNQCKRPRPAAAGSPPRRG 200
FCNQCKRPRPAAAGSPPRRG
Sbjct: 576 FCNQCKRPRPAAAGSPPRRG 635
Score = 90.1 bits (222), Expect(2) = e-146
Identities = 38/38 (100%), Positives = 38/38 (100%)
Frame = +1
Query: 200 GSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGR 237
GSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGR
Sbjct: 634 GSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGR 747
>TC80847 similar to GP|23326831|gb|AAN25338.1 polyribonucleotide
nucleotidyltransferase {Bifidobacterium longum NCC2705},
partial (1%)
Length = 610
Score = 253 bits (647), Expect = 7e-68
Identities = 116/116 (100%), Positives = 116/116 (100%)
Frame = +2
Query: 264 LHRERMDYMDDAYRGRGKFDRPPPLDWDNRDRGRDGFSNERKGFERRPLSPSAPLLPSLP 323
LHRERMDYMDDAYRGRGKFDRPPPLDWDNRDRGRDGFSNERKGFERRPLSPSAPLLPSLP
Sbjct: 2 LHRERMDYMDDAYRGRGKFDRPPPLDWDNRDRGRDGFSNERKGFERRPLSPSAPLLPSLP 181
Query: 324 PHRGGDRWSRDVRDRSRSPIRGGPPAKDYRRDPVMSRGGRDDRRGGVGRDRIGGMY 379
PHRGGDRWSRDVRDRSRSPIRGGPPAKDYRRDPVMSRGGRDDRRGGVGRDRIGGMY
Sbjct: 182 PHRGGDRWSRDVRDRSRSPIRGGPPAKDYRRDPVMSRGGRDDRRGGVGRDRIGGMY 349
>TC82311 weakly similar to PIR|G96696|G96696 protein F1N21.14 [imported] -
Arabidopsis thaliana, partial (23%)
Length = 813
Score = 47.0 bits (110), Expect = 1e-05
Identities = 29/74 (39%), Positives = 35/74 (47%), Gaps = 2/74 (2%)
Frame = +3
Query: 160 REGDWMCPDALCGNLNFARRDFCN--QCKRPRPAAAGSPPRRGSPPLHAPPRRYPGPPFD 217
RE DW CP CGN+NF+ R CN C +PRPA S + PL AP P+
Sbjct: 144 REDDWTCPS--CGNVNFSFRTTCNMRNCTQPRPADHNS--KSAVKPLQAPQAYSSAAPYL 311
Query: 218 RSPERPMNGYRSPP 231
S P +PP
Sbjct: 312 GSNAPPSIYIGAPP 353
Score = 40.8 bits (94), Expect = 9e-04
Identities = 52/197 (26%), Positives = 78/197 (39%), Gaps = 3/197 (1%)
Frame = +3
Query: 2 SSTSPSQSPPSHHHHHQPLLSSLVVRPSLTENSPPGAAAHSNDYEPGELRRDPPPVNYSR 61
S+ P Q+P ++ L S+ PS+ +PP + + P P V ++
Sbjct: 255 SAVKPLQAPQAYSSAAPYLGSN--APPSIYIGAPPYGSLFNGSPVP------PYEVPFTG 410
Query: 62 SDRYSDDAGYRFRAGSS-SPVHHRRDADHRFPSDYNHFPRNRGFGGGRDFGRFRDPPHPY 120
Y G R AGS P+H A + + H GG + D Y
Sbjct: 411 GSAYQYSYGSRVPAGSPYRPLHLAGPAPYTSGAVVGH-------GGIYAMPQLLDR---Y 560
Query: 121 ARGRPGGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNVRPREGDWMCPDALCGNLNFARRD 180
G P G G PG+ R + + + P+ R+ DW CP CGN+NF+ R
Sbjct: 561 GLGVPIGP--GAMGTKPGF-FRDDKSQ----KKGPDAT-RDNDWACPK--CGNVNFSFRT 710
Query: 181 FCN--QCKRPRPAAAGS 195
CN +C P+P + S
Sbjct: 711 VCNMRKCNTPKPGSQAS 761
>BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (39%)
Length = 1358
Score = 45.4 bits (106), Expect = 4e-05
Identities = 76/262 (29%), Positives = 90/262 (34%), Gaps = 15/262 (5%)
Frame = -3
Query: 6 PSQSPPSHHHHHQPLLSS----LVVRPSLTEN---SPPGAA-AHSNDYEPGELRRDPPPV 57
PS++P H P S + RPS EN +PPGA + + Y P PPP
Sbjct: 720 PSRAPRRHPPPPPPPPPSSPPGMGTRPSCQENWWAAPPGARPSRPSRYIPPPT--SPPPA 547
Query: 58 NYSRSDRYSDDAGYRFRAGSSSPVHHRRDADHRFPSDYNHFPRNRGFGGGRDFGRFRDPP 117
+ + R RA +P R A P PR R R P
Sbjct: 546 SPPAPVPPQSPSTIRGRAPPPAPPPRRPPAR---PPPAPQRPRPR-----------RPPA 409
Query: 118 HPYARGRPGGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNVRPREGDWMCPDALCGNLNFA 177
P + R G R R +G G G P+ P P
Sbjct: 408 RPPSGRRRGARAKVRKLP---------KG-GTPGSRAPSPAPPSPTHPAP---------- 289
Query: 178 RRDFCNQCKRPRP-----AAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPP 232
RR + RPRP AA G RG PP H PP P PP P P + RS PP
Sbjct: 288 RRPWTGPPPRPRPHSP*QAACG----RGHPPRHRPPPPGPPPPPPLGPRPPPHPPRSGPP 121
Query: 233 R--LMGRDGLRDYGPAAAALPP 252
R R L P AA PP
Sbjct: 120 RPHPPTRTSLSPPPPPPAASPP 55
Score = 31.6 bits (70), Expect = 0.53
Identities = 44/148 (29%), Positives = 53/148 (35%), Gaps = 11/148 (7%)
Frame = -1
Query: 188 PRPAAAGSPPRRGSPPLHAPPRRY-----PGPPFDRSPERPMNGYRSPPPR---LMGRDG 239
PRP PP SPP APP+ P PP P P+ +P P R G
Sbjct: 563 PRPPPPVPPPP--SPP-RAPPQSEAAPPPPPPPRGARPRAPLRPPSAPDPAGHPRAPRQG 393
Query: 240 LRDY-GPAAAALPPLRHEGRFPDPHLHRERMDYMDDAYRGRGKFDRPPPLDWDNRDRGRD 298
+ GP + P H G P P L R A G G P P ++ +
Sbjct: 392 VAVVPGPRSGNSPRAAHRGPEPRP-LPRPPPPTRPPAAPGPGPPPAPGPTPL-SKPPAAE 219
Query: 299 GF--SNERKGFERRPLSPSAPLLPSLPP 324
G + RR PSAP P PP
Sbjct: 218 GIPPATAPPPRARRLRPPSAPAPPPTPP 135
Score = 31.2 bits (69), Expect = 0.69
Identities = 17/45 (37%), Positives = 19/45 (41%)
Frame = -2
Query: 188 PRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPP 232
PRP PP + PP P P PP P R R+PPP
Sbjct: 166 PRPPPPPPPPPQRPPPPPPPHTHQPEPP-TSPPRRQPPAPRAPPP 35
Score = 30.4 bits (67), Expect = 1.2
Identities = 33/107 (30%), Positives = 41/107 (37%), Gaps = 7/107 (6%)
Frame = -2
Query: 155 PNVRPREGDW-MCPDALCGNLNFARRDFCNQCKRPRPAAAGSP-----PRRGSPPLHAP- 207
P + PR G+ + P L G+ ARR + PA P PR PLH P
Sbjct: 673 PLLPPRNGNASLLPRKLVGSP--ARRSSLSSESLHTPAHLAPPRQSPRPRPPPEPLHNPR 500
Query: 208 PRRYPGPPFDRSPERPMNGYRSPPPRLMGRDGLRDYGPAAAALPPLR 254
PR P PP P +G +PP P A PP+R
Sbjct: 499 PRPPPRPPPAAPARAPPSGPPAPPT------------PPATRAPPVR 395
Score = 28.5 bits (62), Expect = 4.5
Identities = 36/113 (31%), Positives = 40/113 (34%), Gaps = 9/113 (7%)
Frame = -1
Query: 159 PREGDWMCPDALCGNLNFA--RRDFCNQCKRP----RPAAA---GSPPRRGSPPLHAPPR 209
PR+G + P GN A R RP RP AA G PP G PL PP
Sbjct: 404 PRQGVAVVPGPRSGNSPRAAHRGPEPRPLPRPPPPTRPPAAPGPGPPPAPGPTPLSKPPA 225
Query: 210 RYPGPPFDRSPERPMNGYRSPPPRLMGRDGLRDYGPAAAALPPLRHEGRFPDP 262
PP +PPPR R P+A A PP P P
Sbjct: 224 AEGIPP-----------ATAPPPR-----ARRLRPPSAPAPPPTPPAAAPPAP 114
Score = 28.1 bits (61), Expect = 5.8
Identities = 24/69 (34%), Positives = 28/69 (39%), Gaps = 3/69 (4%)
Frame = -2
Query: 187 RPRPAAAGSPPRRGSPPLHAPPRR---YPGPPFDRSPERPMNGYRSPPPRLMGRDGLRDY 243
RPR + PP G P APPR P PP P P + ++ PP R
Sbjct: 226 RPRASPPPPPPPPG-PAASAPPRPPPPPPPPPQRPPPPPPPHTHQPEPP----TSPPRRQ 62
Query: 244 GPAAAALPP 252
PA A PP
Sbjct: 61 PPAPRAPPP 35
Score = 28.1 bits (61), Expect = 5.8
Identities = 21/51 (41%), Positives = 22/51 (42%)
Frame = +3
Query: 99 PRNRGFGGGRDFGRFRDPPHPYARGRPGGRPFGRAFDGPGYGHRHGRGEGL 149
P G GGG GR R A G G R R GPG R G+G GL
Sbjct: 186 PGGGGGGGGDALGRRRLAKGSGAGGGGGAR--SRGGGGPGGWGRAGQGTGL 332
Score = 27.3 bits (59), Expect = 9.9
Identities = 25/87 (28%), Positives = 28/87 (31%), Gaps = 6/87 (6%)
Frame = -3
Query: 188 PRPAAAGSPPRRGSPPL------HAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDGLR 241
P P SPP G+ P APP P P P SPP + +
Sbjct: 687 PPPPPPSSPPGMGTRPSCQENWWAAPPGARPSRPSRYIPPPTSPPPASPPAPVPPQSPST 508
Query: 242 DYGPAAAALPPLRHEGRFPDPHLHRER 268
G A PP R P P R R
Sbjct: 507 IRGRAPPPAPPPRRPPARPPPAPQRPR 427
Score = 27.3 bits (59), Expect = 9.9
Identities = 27/87 (31%), Positives = 30/87 (34%), Gaps = 8/87 (9%)
Frame = -2
Query: 188 PRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDGLRDYGPAA 247
P P A P PPL P P P S RP PPP GPAA
Sbjct: 328 PVPCPALPHPPGPPPPLDRAPPPPPAPLPLASRLRPRASPPPPPP---------PPGPAA 176
Query: 248 AA--------LPPLRHEGRFPDPHLHR 266
+A PP + P PH H+
Sbjct: 175 SAPPRPPPPPPPPPQRPPPPPPPHTHQ 95
>BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xylostella
granulovirus}, partial (42%)
Length = 909
Score = 43.5 bits (101), Expect = 1e-04
Identities = 48/175 (27%), Positives = 64/175 (36%), Gaps = 13/175 (7%)
Frame = +2
Query: 186 KRPRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRS--PERPMNGYRSPPPRLMGRDGLRDY 243
+R RP G PPRRG P P PP R +P G + R G++G +
Sbjct: 344 RRRRPNKPGPPPRRGGGQGGRAPAPPPPPPKAREGRTPKPDPGSKENHNRTPGKEGPATH 523
Query: 244 GPAAAALPPL------RHEGRF-----PDPHLHRERMDYMDDAYRGRGKFDRPPPLDWDN 292
GP PP +H R P PH R G G PPP N
Sbjct: 524 GPPEKKTPPPTPKRRGKHTKRAPGGGPPPPHPGR----------GGGGPGGPPPPPPKKN 673
Query: 293 RDRGRDGFSNERKGFERRPLSPSAPLLPSLPPHRGGDRWSRDVRDRSRSPIRGGP 347
+RG R G +R+ A P P + R +R+ R + + P + P
Sbjct: 674 SERGGRAAPRGRPGTKRKNPRRPAGSGPRGAPAKKTPRRTRE-RGKKKKPPKKTP 835
Score = 35.4 bits (80), Expect = 0.037
Identities = 50/212 (23%), Positives = 69/212 (31%), Gaps = 7/212 (3%)
Frame = +3
Query: 28 PSLTENSPPGA--AAHSNDYEPGEL-----RRDPPPVNYSRSDRYSDDAGYRFRAGSSSP 80
P T +PP A A S + PGE PPP R R R +++
Sbjct: 315 PGTTRGTPPTADDAQTSPAHHPGEGGGKGGAPPPPPRPPPRHGRGGHQNQTREAKRTTTE 494
Query: 81 VHHRRDADHRFPSDYNHFPRNRGFGGGRDFGRFRDPPHPYARGRPGGRPFGRAFDGPGYG 140
+RD P H P ++ GG + P P+ GG P G P
Sbjct: 495 PQGKRDRPRTDPRKKKHPPPHQKEGGNTQKEPRGEDPPPHTPAGGGGGPGGPRPPPPKKT 674
Query: 141 HRHGRGEGLHGRNNPNVRPREGDWMCPDALCGNLNFARRDFCNQCKRPRPAAAGSPPRRG 200
R G G + R GD + RPR G+ R G
Sbjct: 675 ARGGEERPPAGARARREKTR-GD---------------QQEAAPAGRPRKKPRGAREREG 806
Query: 201 SPPLHAPPRRYPGPPFDRSPERPMNGYRSPPP 232
P++ P P + P+ P ++PPP
Sbjct: 807 K---KKNPQKKPPKPQQQPPKNPPPTKKTPPP 893
Score = 35.0 bits (79), Expect = 0.048
Identities = 32/123 (26%), Positives = 39/123 (31%), Gaps = 19/123 (15%)
Frame = +1
Query: 184 QCKRPRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERP---------MNGYRSPPPRL 234
Q + P+P A +R PPR P PP P+ P PPP
Sbjct: 58 QARNPQPEEATKKQKRAPEKKKNPPRGQPPPPPPPPPQAPPTKPPTRQTPKNNTPPPPPH 237
Query: 235 MGRDGLRDYGPAAAALPPLRHE----------GRFPDPHLHRERMDYMDDAYRGRGKFDR 284
D P PP HE GR P P ++ + RG R
Sbjct: 238 TPPDPTPPQPPPQPPKPP-HHEKRPRTPEPPGGRPPPPTTPKQARPTTQERGGARGARPR 414
Query: 285 PPP 287
PPP
Sbjct: 415 PPP 423
Score = 34.7 bits (78), Expect = 0.062
Identities = 51/204 (25%), Positives = 70/204 (34%), Gaps = 22/204 (10%)
Frame = +3
Query: 183 NQCKRPRPAAAGSPPRRGSPPL------HAPPRRYP-GPPFDRSPERPMNGYRSPPPRLM 235
N K+ P PPR +PP PPR P P R + ++ P
Sbjct: 201 NPKKQHTPTTPTHPPRPHTPPTPPPTTQTTPPREAPTDPGTTRGTPPTADDAQTSPAHHP 380
Query: 236 GRDGLRDYGPAAAALPPLRH-EGRFPDPHLHRERMDYMDDAYRGRGKFD-----RPPPL- 288
G G + P PP RH G + +R R R + D PPP
Sbjct: 381 GEGGGKGGAPPPPPRPPPRHGRGGHQNQTREAKRTTTEPQGKRDRPRTDPRKKKHPPPHQ 560
Query: 289 ----DWDNRDRGRDGFSNERKGFERRPLSPSAPLLPSLPPHRGGDR----WSRDVRDRSR 340
+ RG D + G P P P P RGG+ +R R+++R
Sbjct: 561 KEGGNTQKEPRGEDPPPHTPAGGGGGPGGPRPP--PPKKTARGGEERPPAGARARREKTR 734
Query: 341 SPIRGGPPAKDYRRDPVMSRGGRD 364
+ PA R+ P RG R+
Sbjct: 735 GDQQEAAPAGRPRKKP---RGARE 797
Score = 33.5 bits (75), Expect = 0.14
Identities = 55/241 (22%), Positives = 73/241 (29%), Gaps = 60/241 (24%)
Frame = +3
Query: 188 PRPAAAGSPPRRGSPPLHAP-----------PRRYPGPPFDRSPERPMNGYRSPPPRLMG 236
P P+ +P R+G P P PR+ PP + P PP +
Sbjct: 15 PTPSPQATPQRKGHPSQKPPTGRSHKKTKKSPRKKEKPPPRPATPPPAPAPPGPPNQTTN 194
Query: 237 -RDGLRDYGPAAAALPPLRHEGRFP--------------DPHLHRERMDYMDDAY----- 276
+ + + P PP H P DP R DDA
Sbjct: 195 TTNPKKQHTPTTPTHPPRPHTPPTPPPTTQTTPPREAPTDPGTTRGTPPTADDAQTSPAH 374
Query: 277 ---RGRGKFDRPPPLDWDNRDRGRDGFSNERKGFERRPLSPSA--------PLLPSLPP- 324
G GK PPP GR G N+ + +R P P PP
Sbjct: 375 HPGEGGGKGGAPPPPPRPPPRHGRGGHQNQTREAKRTTTEPQGKRDRPRTDPRKKKHPPP 554
Query: 325 -HRGGDRWSRDVRDRS---RSPIRGG---------PPAKDYR----RDPVMSRGGRDDRR 367
+ G ++ R +P GG PP K R R P +R R+ R
Sbjct: 555 HQKEGGNTQKEPRGEDPPPHTPAGGGGGPGGPRPPPPKKTARGGEERPPAGARARREKTR 734
Query: 368 G 368
G
Sbjct: 735 G 737
Score = 32.3 bits (72), Expect = 0.31
Identities = 41/191 (21%), Positives = 61/191 (31%), Gaps = 5/191 (2%)
Frame = +2
Query: 188 PRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDGLRDYGPAA 247
P P PP++ +P P R+ +P + PP + R P A
Sbjct: 14 PNPQPPSDPPKKRTPKPETPNRK-----------KPQKNKKEPPKK-------RKTPPEA 139
Query: 248 AALPPLRHEGRFPDPHLHRERMDYMDDAYRGRGKFDRP----PPLDWDNRDRGRDGFSNE 303
+ PP R P P+ ++ + P PP + N R
Sbjct: 140 SHPPPRPRPPRPPQPNHQHDKPQKTTHPHHPHTPPPTPHPPNPPPNHPNHPTTRSA-HGP 316
Query: 304 RKGFERRPLSPSAPLLPSLPPHRGGDRWSRDVRDRSRSPIRGGPPAKDYR-RDPVMSRGG 362
R P P P PP RGG + R+P PP K R P G
Sbjct: 317 RNHPGDAPHRRRRPNKPGPPPRRGGGQ-------GGRAPAPPPPPPKAREGRTPKPDPGS 475
Query: 363 RDDRRGGVGRD 373
+++ G++
Sbjct: 476 KENHNRTPGKE 508
Score = 31.6 bits (70), Expect = 0.53
Identities = 58/247 (23%), Positives = 71/247 (28%), Gaps = 58/247 (23%)
Frame = +2
Query: 186 KRPRPAAAGSPPRRGSPP--LHAPPR---------------------------------- 209
K+P+ P +R +PP H PPR
Sbjct: 80 KKPQKNKKEPPKKRKTPPEASHPPPRPRPPRPPQPNHQHDKPQKTTHPHHPHTPPPTPHP 259
Query: 210 -----RYPGPPFDRSPERPMN------------GYRSPPPRLMGRDGLRDYGPAAAALPP 252
+P P RS P N PPPR G G R PA PP
Sbjct: 260 PNPPPNHPNHPTTRSAHGPRNHPGDAPHRRRRPNKPGPPPRRGGGQGGR--APAPPPPPP 433
Query: 253 LRHEGRFPDP-----HLHRERMDYMDDAYRGRGKFDRPPPLDWDNRDRGRDGFSNERKGF 307
EGR P P H A G + PPP + +R+G
Sbjct: 434 KAREGRTPKPDPGSKENHNRTPGKEGPATHGPPEKKTPPP-------------TPKRRGK 574
Query: 308 ERRPLSPSAPLLPSLPPHRGGDRWSRDVRDRSRSPIRGGPPAKDYRRDPVMSRGGRDDRR 367
+ P P P RGG GGPP +++ RGGR R
Sbjct: 575 HTKRAPGGGP--PPPHPGRGGGG-------------PGGPPPPPPKKN--SERGGRAAPR 703
Query: 368 GGVGRDR 374
G G R
Sbjct: 704 GRPGTKR 724
>TC88921 similar to SP|P92966|RS41_ARATH Arginine/serine-rich splicing
factor RSP41. [Mouse-ear cress] {Arabidopsis thaliana},
partial (33%)
Length = 1419
Score = 43.1 bits (100), Expect = 2e-04
Identities = 64/244 (26%), Positives = 83/244 (33%), Gaps = 14/244 (5%)
Frame = +1
Query: 52 RDPPPVNYSRSDRYSDDAGYRFRAGSSSPVHHRRDADHRFPSDYNHFPRNRGFGGGRDFG 111
RD N DR D R R G SP +RR+ G D+G
Sbjct: 292 RDDDRRNGHSPDRGRDRQRDRSRDGRRSPSPYRRER------------------GSPDYG 417
Query: 112 RFRDPPHPYARGRPGGRPFGRAFDGPGYGHRHGRGEGLHGRNN--PNVRPREGDWMCPDA 169
R P PY R R +GR HR RG +GR N PN R R+G D
Sbjct: 418 R---GPSPYKRER-SSPDYGRGNSRSRSPHRRERGSPAYGRRNPSPNRRERDGAEAVRDT 585
Query: 170 LCGNLNFARRDFCNQCKRPRPAAAGSPPRRG---SPPLHAPPRRYPGPPFD--RSPE--- 221
+ RR P++G P + + P D RSP+
Sbjct: 586 SRSPYHKERRRTDRSRSHSLEEGERIEPQKGHGSDPSPYGTVKDSPENGHDRRRSPDAKR 765
Query: 222 --RPMNGYR-SPPPRLMGRDG-LRDYGPAAAALPPLRHEGRFPDPHLHRERMDYMDDAYR 277
P NG R SP P G DYG + +P R + P M++ YR
Sbjct: 766 NPSPFNGSRGSPSPDAKGNPSPYNDYGGSPNTMPEPRDSPNYGGPE------SPMNEQYR 927
Query: 278 GRGK 281
+ +
Sbjct: 928 SQSQ 939
Score = 31.6 bits (70), Expect = 0.53
Identities = 44/157 (28%), Positives = 54/157 (34%)
Frame = +1
Query: 217 DRSPERPMNGYRSPPPRLMGRDGLRDYGPAAAALPPLRHEGRFPDPHLHRERMDYMDDAY 276
DR +R +G RSP P R G DYG R P P+ RER D
Sbjct: 337 DRQRDRSRDGRRSPSPYRRER-GSPDYG-------------RGPSPY-KRERSS--PDYG 465
Query: 277 RGRGKFDRPPPLDWDNRDRGRDGFSNERKGFERRPLSPSAPLLPSLPPHRGGDRWSRDVR 336
RG + P R+RG + RR R G + VR
Sbjct: 466 RGNSRSRSP-----HRRERGSPAYGRRNPSPNRR--------------ERDG---AEAVR 579
Query: 337 DRSRSPIRGGPPAKDYRRDPVMSRGGRDDRRGGVGRD 373
D SRSP D R + G R + + G G D
Sbjct: 580 DTSRSPYHKERRRTDRSRSHSLEEGERIEPQKGHGSD 690
>BQ141681 weakly similar to GP|15145793|gb| basic proline-rich protein {Sus
scrofa}, partial (9%)
Length = 516
Score = 42.4 bits (98), Expect = 3e-04
Identities = 48/163 (29%), Positives = 60/163 (36%), Gaps = 10/163 (6%)
Frame = +1
Query: 114 RDPPH-PYARGRPGGRPFGRAFD-GPGYGHRHGRGEGLHGRNNPNVRPREGDWMCPDALC 171
R PPH P RG+P + R GP G + +G+ +N P R R
Sbjct: 109 RGPPHKPKTRGQPNQKQEHRPKTRGPNKGPKP-KGKNNPQKNGPRKRRRT---------- 255
Query: 172 GNLNFARRDFCNQCKRPRPAAAGSP-------PRRGSPPLHAPPRRYPGPPFDRSPERPM 224
ARR + +R +P +G+P PR+ PP PRR P P P RP
Sbjct: 256 -----ARRRSSARGRRGKPGESGTPTPPGTAAPRKRPPPQGRAPRRAP-PGGPGGPARPP 417
Query: 225 NGYRSPPPRLMGRDGLRDYGPAAAALPPLRHEG-RFPDPHLHR 266
PPP PA P R G R P P R
Sbjct: 418 PDAARPPP------------PAGPGANPARRAGRRGPAPRARR 510
Score = 29.3 bits (64), Expect = 2.6
Identities = 31/96 (32%), Positives = 39/96 (40%), Gaps = 7/96 (7%)
Frame = +2
Query: 279 RGKFDRPPPLDWDNRDRGRDGFSNERKGFE-----RRPLSPSAPLL--PSLPPHRGGDRW 331
RGK + PP R D + E+ G + RRP P P+ PP D
Sbjct: 236 RGKGEEPPE---GEAARAGDEENREKAGPQPPRGPRRPEKDPPPRAEPPAAPPP--ADPE 400
Query: 332 SRDVRDRSRSPIRGGPPAKDYRRDPVMSRGGRDDRR 367
R R+R RG PP + R P +RGG RR
Sbjct: 401 GRPAPPRTR---RGPPPPRARARTPPGARGGGGRRR 499
>TC90113 similar to PIR|C86301|C86301 arginine/serine-rich protein
[imported] - Arabidopsis thaliana, partial (70%)
Length = 1259
Score = 42.4 bits (98), Expect = 3e-04
Identities = 59/192 (30%), Positives = 65/192 (33%), Gaps = 5/192 (2%)
Frame = +1
Query: 188 PRPAAAGSPPRRGSP--PLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDGLRDYGP 245
PR AG+ + P P + PRR P P SP R RSP PR G R P
Sbjct: 553 PRTDNAGAEVEKDGPKRPRESSPRRKPPP----SPRR-----RSPVPRRAGSPR-RPESP 702
Query: 246 AAAALPPLRHEGRFPDPHLHRERMDYMDDAYRGRGKFDRPPPLDWDNRDRGRDGFSNERK 305
A P+R +D YR RG D PP
Sbjct: 703 RRRADSPVRRR---------------LDSPYR-RGAGDTPP------------------- 777
Query: 306 GFERRPLSPSAPLLPSLPPHRGGDRWSRDVRDRSRSP---IRGGPPAKDYRRDPVMSRGG 362
RRP+SP PS PP R R SP R PP RR P R
Sbjct: 778 --RRRPVSPGRGRSPSPPPRRLRSPARVSPRRMRGSPGPGRRRSPPPPPRRRSP-PRRAR 948
Query: 363 RDDRRGGVGRDR 374
RR VGR R
Sbjct: 949 SPPRRSPVGRRR 984
Score = 37.4 bits (85), Expect = 0.010
Identities = 56/198 (28%), Positives = 70/198 (35%), Gaps = 16/198 (8%)
Frame = +1
Query: 51 RRDPPPVNYSRSDRYSDDAGYRF----RAGSSSPVHHRRDADHRFPSDYNHFPRNRGFGG 106
RR PPP RS R R + SPV R D+ +R + PR R
Sbjct: 622 RRKPPPSPRRRSPVPRRAGSPRRPESPRRRADSPVRRRLDSPYRRGAGDTP-PRRRPVSP 798
Query: 107 GRDFGRFRDPPHPYARGRPGGRPFGRAFDG-PGYGHRHGRGEGLHGRNNPNVRPREGDWM 165
GR R P P R R R R G PG G R R+ P R R
Sbjct: 799 GRG----RSPSPPPRRLRSPARVSPRRMRGSPGPGRRRSPPPPPRRRSPPR-RARSPPRR 963
Query: 166 CPDALCGNLNFARRDFCNQCKRPRPAAAGSPPRRGSPPLHAP---PRRYPGPPFDRSPER 222
P + + RR ++ + P P RRG ++ PR+ RSP R
Sbjct: 964 SPVGRRRSRSPIRRSARSRSRSFSPRRGRPPMRRGRSSSYSDSPSPRKVSRRSKSRSPRR 1143
Query: 223 PMNG--------YRSPPP 232
P+ G SPPP
Sbjct: 1144PLKGRASSNSSSSSSPPP 1197
Score = 32.0 bits (71), Expect = 0.40
Identities = 24/56 (42%), Positives = 25/56 (43%), Gaps = 9/56 (16%)
Frame = +1
Query: 187 RPRPAAAG------SPPRRGSPPLHAPPRRY---PGPPFDRSPERPMNGYRSPPPR 233
R RP + G PPRR P PRR PGP RSP P RSPP R
Sbjct: 778 RRRPVSPGRGRSPSPPPRRLRSPARVSPRRMRGSPGPGRRRSPPPPPR-RRSPPRR 942
Score = 29.6 bits (65), Expect = 2.0
Identities = 17/38 (44%), Positives = 22/38 (57%), Gaps = 2/38 (5%)
Frame = +1
Query: 188 PRPAAAGSPPRRG-SPPLHAPPR-RYPGPPFDRSPERP 223
PR ++ P RRG SPPL PP+ + P PP P +P
Sbjct: 142 PRRKSSAEPARRGRSPPLPPPPQSKRPSPP----PRKP 243
>TC81177 similar to GP|15293081|gb|AAK93651.1 unknown protein {Arabidopsis
thaliana}, partial (28%)
Length = 632
Score = 42.0 bits (97), Expect = 4e-04
Identities = 49/158 (31%), Positives = 59/158 (37%), Gaps = 2/158 (1%)
Frame = +1
Query: 187 RPRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDGLRDYGPA 246
R R + S RR P PR PP RS R Y SPPP+ R+Y +
Sbjct: 253 RERSSGRHSDRRRSPPRYSRSPRYSRSPPRHRSRSRGSRDYHSPPPK------RREYSRS 414
Query: 247 AAALPPLRHEGRFPDPHLHRERMDYMDDAYRGRGKFDRPPPLDWDNRDRGRDGFSNERKG 306
A+ P R R H RER ++ R PP + D R R + S +
Sbjct: 415 AS--PEDRRHSREGSQH-SRER------------EYSRSPPKNGDARSRSQ---SPVKGS 540
Query: 307 FERRPLSPSAPLLPSLPPHRGGDRWSRDVR--DRSRSP 342
E R SP SRDVR RSRSP
Sbjct: 541 VESRSPSP-----------------SRDVRAASRSRSP 603
>TC76916 MtN4
Length = 1850
Score = 41.2 bits (95), Expect = 7e-04
Identities = 25/84 (29%), Positives = 29/84 (33%), Gaps = 2/84 (2%)
Frame = +1
Query: 182 CNQCKRPRPAAAGSPPRRGSPPLHAPPRR--YPGPPFDRSPERPMNGYRSPPPRLMGRDG 239
C C P P P PP+H PP+ P PP + P P PPP +
Sbjct: 496 CPYCPYPSPKPPSHHPPIVKPPVHKPPKHSPTPKPPVHKPPRYPPKPSPCPPPSSTPK-- 669
Query: 240 LRDYGPAAAALPPLRHEGRFPDPH 263
P PP H P PH
Sbjct: 670 -----PPHHPKPPAVHPPHVPKPH 726
>TC82284 weakly similar to GP|11119510|gb|AAF37724.2 diaphanous protein
{Entamoeba histolytica}, partial (4%)
Length = 671
Score = 40.8 bits (94), Expect = 9e-04
Identities = 27/67 (40%), Positives = 31/67 (45%), Gaps = 5/67 (7%)
Frame = +1
Query: 192 AAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPM-NGYRSPPPRLMGR----DGLRDYGPA 246
A G PPR PP AP P PP P PM N R+PPP + G + GP
Sbjct: 40 APGVPPRPMPPPPQAPLPPPPPPPQGLPPGAPMGNPPRAPPPPMSGSMPPPPPMAANGPR 219
Query: 247 AAALPPL 253
A+PPL
Sbjct: 220 PGAMPPL 240
Score = 27.3 bits (59), Expect = 9.9
Identities = 17/49 (34%), Positives = 21/49 (42%)
Frame = +1
Query: 188 PRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMG 236
P A G+PPR PP+ P PP + RP PPP +G
Sbjct: 121 PPGAPMGNPPRAPPPPMSG--SMPPPPPMAANGPRPGAMPPLPPPAPIG 261
>TC92146 weakly similar to PIR|T37870|T37870 RNA-binding / Ran zinc finger
proteinrotein - fission yeast (Schizosaccharomyces
pombe), partial (4%)
Length = 459
Score = 40.8 bits (94), Expect = 9e-04
Identities = 16/39 (41%), Positives = 24/39 (61%)
Frame = +2
Query: 156 NVRPREGDWMCPDALCGNLNFARRDFCNQCKRPRPAAAG 194
N++P GDW+C + +CG NFA+R C +C P + G
Sbjct: 80 NMKP--GDWLCGNEMCGYHNFAKRTHCAKCGMPNNSTVG 190
>AJ498860 similar to GP|20197423|gb|A unknown protein {Arabidopsis thaliana},
partial (80%)
Length = 596
Score = 40.4 bits (93), Expect = 0.001
Identities = 27/82 (32%), Positives = 33/82 (39%), Gaps = 4/82 (4%)
Frame = +1
Query: 188 PRPAAAGSPPR----RGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDGLRDY 243
P A G PP+ +G PP PP+ YP P + P GY PPP + Y
Sbjct: 79 PSDAKEGHPPQGYPPQGYPPQGYPPQGYPPPGYPPPGYPPQQGY--PPPPYPAQGYPPQY 252
Query: 244 GPAAAALPPLRHEGRFPDPHLH 265
P A PP P H+H
Sbjct: 253 APPQYAQPP-------PPQHVH 297
>TC91856 similar to GP|11994340|dbj|BAB02299. zinc finger protein-like;
Ser/Thr protein kinase-like protein {Arabidopsis
thaliana}, partial (64%)
Length = 663
Score = 38.9 bits (89), Expect = 0.003
Identities = 20/54 (37%), Positives = 28/54 (51%), Gaps = 2/54 (3%)
Frame = +3
Query: 143 HGRGEGLHGRNNPNVRP--REGDWMCPDALCGNLNFARRDFCNQCKRPRPAAAG 194
+G G G G + RP + GDW+C + C NFA R C +C PR ++ G
Sbjct: 414 YGFGAGSAG-GGASTRPGWKSGDWICTRSGCNEHNFANRMECYRCNGPRDSSTG 572
Score = 38.5 bits (88), Expect = 0.004
Identities = 29/85 (34%), Positives = 33/85 (38%), Gaps = 2/85 (2%)
Frame = +3
Query: 126 GGRPFGRAFDGPGYGHRHGRGEG--LHGRNNPNVRPREGDWMCPDALCGNLNFARRDFCN 183
G FG +F G GRG P+VRP GDW C CG NFA R C
Sbjct: 186 GAVDFGGSFLG-------GRGSSSPFPFTTGPDVRP--GDWYCTVGNCGAHNFASRSSCF 338
Query: 184 QCKRPRPAAAGSPPRRGSPPLHAPP 208
+C P+ S P L P
Sbjct: 339 KCGAPKDIDTFSSDSSDMPRLLRSP 413
>BQ137255 homologue to PIR|B34768|B347 ORF5 protein - Orf virus (strain NZ2),
partial (10%)
Length = 1161
Score = 38.9 bits (89), Expect = 0.003
Identities = 47/154 (30%), Positives = 51/154 (32%), Gaps = 26/154 (16%)
Frame = +2
Query: 190 PAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPE-RPMN-----GYRSPPPRLMGRDGLRDY 243
P S + PP H PP R P PP R RP G R P RDG
Sbjct: 206 PQTHHSRQAKRRPPHHTPPPRNPRPPRPRGRRPRPHGGPTAAGARGGPRSSARRDGRAMG 385
Query: 244 GPA--AAALPPLRHEGRFPDP----------------HLHRERMDYMDDAYRGRGKFDRP 285
PA A A PP R + P HR R R R + R
Sbjct: 386 APAVVARAPPPCRRNDQTKRPTE*RARSERASERRERAQHRARRTTRARVARPRARDARA 565
Query: 286 PPLDWDNRDRGRDGFSNERKGFE--RRPLSPSAP 317
D R RGR S R E RRP +P P
Sbjct: 566 TRARNDERARGRVARSRRRTTRERRRRPTAPPTP 667
Score = 30.4 bits (67), Expect = 1.2
Identities = 47/184 (25%), Positives = 59/184 (31%), Gaps = 15/184 (8%)
Frame = +1
Query: 198 RRGSPPLHAPPRRYPGPPFDRS----PERPMNGYRSPPPRLMGRDGLR-------DYGPA 246
+ G+PP PP P +R P RP N R+ PP DG+R D
Sbjct: 49 KAGAPPPGPPPSNRRSPQTERRRNERPRRPANATRAAPPG-ENADGIRRTPQNPTDPSQP 225
Query: 247 AAALPPLRHEGRFPDPHLHRERMDYMDDAYRGRGKFDRPPPLDWDNRDRGRDGFSNERKG 306
+ PP P A R G P R R G S
Sbjct: 226 TSQAPPAPPHPAASQPTTPPSARPAPAPARRPDGGGGARRPAIERAARRTRHGSSGGG-- 399
Query: 307 FERRPLSPSAPLLPSLPPHRGGDRWSRDVRDRSR----SPIRGGPPAKDYRRDPVMSRGG 362
PSA +P P++ DR +R R R R S + P RR +R
Sbjct: 400 ------GPSASAVPQERPNQATDRVARAERARVRAARASATQSAPNDARARRASARTRCA 561
Query: 363 RDDR 366
D R
Sbjct: 562 GDAR 573
>TC83142 similar to GP|20197597|gb|AAM15145.1 predicted protein {Arabidopsis
thaliana}, partial (57%)
Length = 865
Score = 38.9 bits (89), Expect = 0.003
Identities = 16/29 (55%), Positives = 19/29 (65%)
Frame = +3
Query: 161 EGDWMCPDALCGNLNFARRDFCNQCKRPR 189
E DW C C N N+A R FCN+CK+PR
Sbjct: 21 ERDWECSG--CNNRNYAFRSFCNRCKQPR 101
Score = 37.7 bits (86), Expect = 0.007
Identities = 26/79 (32%), Positives = 34/79 (42%), Gaps = 4/79 (5%)
Frame = +3
Query: 160 REGDWMCPDALCGNLNFARRDFCNQCKRPRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRS 219
R GDW+C C N N+A R+ C +C +P+ AA A P + P
Sbjct: 147 RIGDWICTG--CTNNNYASREKCKKCGQPKEVAAMPAIAMTGASFSAYPHYFSRVP--GG 314
Query: 220 PERPMN----GYRSPPPRL 234
PE+ MN G PP L
Sbjct: 315 PEQRMNIGLIGNGGPPQSL 371
>AL375750 weakly similar to PIR|A96592|A965 hypothetical protein F14C21.55
[imported] - Arabidopsis thaliana, partial (3%)
Length = 528
Score = 37.7 bits (86), Expect = 0.007
Identities = 21/61 (34%), Positives = 30/61 (48%), Gaps = 2/61 (3%)
Frame = +2
Query: 155 PNVRPRE--GDWMCPDALCGNLNFARRDFCNQCKRPRPAAAGSPPRRGSPPLHAPPRRYP 212
P RP++ GDW CP C +NFA +D C +C+ P P +P ++YP
Sbjct: 20 PEPRPKKHPGDWSCPK--CDFMNFASKDKCFRCQESNPNPNKYPGEWPNP----NSKKYP 181
Query: 213 G 213
G
Sbjct: 182 G 184
Score = 33.1 bits (74), Expect = 0.18
Identities = 14/32 (43%), Positives = 17/32 (52%)
Frame = +2
Query: 154 NPNVRPREGDWMCPDALCGNLNFARRDFCNQC 185
NPN + GDW CP C N+AR C +C
Sbjct: 158 NPNSKKYPGDWSCPK--CDFYNYARNTTCLKC 247
>TC76716 similar to PIR|T10863|T10863 extensin precursor - kidney bean,
partial (48%)
Length = 1275
Score = 37.7 bits (86), Expect = 0.007
Identities = 22/58 (37%), Positives = 28/58 (47%), Gaps = 11/58 (18%)
Frame = +3
Query: 186 KRPRPAAAGSPP-----RRGSPPLHAPPRRY-----PGPPFDR-SPERPMNGYRSPPP 232
K P P PP + PP+H+PP Y P PP+ SP P+ Y+SPPP
Sbjct: 459 KSPPPPVYSPPPPVYKYKSPPPPVHSPPPPYKYKSPPPPPYKYPSPPPPVYKYKSPPP 632
Score = 37.4 bits (85), Expect = 0.010
Identities = 22/53 (41%), Positives = 27/53 (50%), Gaps = 6/53 (11%)
Frame = +3
Query: 186 KRPRPAAAGSPP--RRGSPPLHAPPRRYPGPPFD----RSPERPMNGYRSPPP 232
K P P PP + SPP PP +YP PP +SP P+ Y+SPPP
Sbjct: 252 KSPPPPVYSPPPPYKYKSPP--PPPYKYPSPPPPVYKYKSPPPPVYKYKSPPP 404
Score = 37.4 bits (85), Expect = 0.010
Identities = 22/53 (41%), Positives = 27/53 (50%), Gaps = 6/53 (11%)
Frame = +3
Query: 186 KRPRPAAAGSPP--RRGSPPLHAPPRRYPGPPFD----RSPERPMNGYRSPPP 232
K P P PP + SPP PP +YP PP +SP P+ Y+SPPP
Sbjct: 510 KSPPPPVHSPPPPYKYKSPP--PPPYKYPSPPPPVYKYKSPPPPVYKYKSPPP 662
Score = 35.8 bits (81), Expect = 0.028
Identities = 21/57 (36%), Positives = 28/57 (48%), Gaps = 10/57 (17%)
Frame = +3
Query: 186 KRPRPAAAGSPP----RRGSPPLHAPPRRY-----PGPPFDR-SPERPMNGYRSPPP 232
K P P PP + PP+++PP Y P PP+ SP P+ Y+SPPP
Sbjct: 204 KSPPPPVYSPPPVYKYKSPPPPVYSPPPPYKYKSPPPPPYKYPSPPPPVYKYKSPPP 374
Score = 33.5 bits (75), Expect = 0.14
Identities = 25/89 (28%), Positives = 35/89 (39%)
Frame = +3
Query: 199 RGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDGLRDYGPAAAALPPLRHEGR 258
+ PP P +YP PP P+ Y+SPPP + Y P PP ++ +
Sbjct: 390 KSPPPPPKKPYKYPSPP------PPVYKYKSPPPPV--------YSP-----PPPVYKYK 512
Query: 259 FPDPHLHRERMDYMDDAYRGRGKFDRPPP 287
P P +H Y K+ PPP
Sbjct: 513 SPPPPVHSPPPPY---------KYKSPPP 572
Score = 32.3 bits (72), Expect = 0.31
Identities = 19/69 (27%), Positives = 30/69 (42%), Gaps = 4/69 (5%)
Frame = +3
Query: 199 RGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDGLRDY----GPAAAALPPLR 254
+ PP P +YP PP P+ Y+SPPP + + Y P + PP +
Sbjct: 135 KSPPPPPKKPYKYPSPP------PPVYKYKSPPPPVYSPPPVYKYKSPPPPVYSPPPPYK 296
Query: 255 HEGRFPDPH 263
++ P P+
Sbjct: 297 YKSPPPPPY 323
Score = 32.0 bits (71), Expect = 0.40
Identities = 15/38 (39%), Positives = 20/38 (52%), Gaps = 4/38 (10%)
Frame = +3
Query: 199 RGSPPLHAPPRRYPGPPFD----RSPERPMNGYRSPPP 232
+ PP P +YP PP +SP P+ Y+SPPP
Sbjct: 648 KSPPPPPKKPYKYPSPPPPVYKYKSPPPPVYKYKSPPP 761
>BQ141029 similar to GP|8132441|gb| extensin {Pisum sativum}, partial (54%)
Length = 613
Score = 37.4 bits (85), Expect = 0.010
Identities = 22/53 (41%), Positives = 27/53 (50%), Gaps = 6/53 (11%)
Frame = +2
Query: 186 KRPRPAAAGSPP--RRGSPPLHAPPRRYPGPPFD----RSPERPMNGYRSPPP 232
K P P PP + SPP PP +YP PP +SP P+ Y+SPPP
Sbjct: 275 KSPPPPVYSPPPPYKYKSPP--PPPYKYPSPPPPVYKYKSPPPPVYKYKSPPP 427
Score = 35.8 bits (81), Expect = 0.028
Identities = 21/57 (36%), Positives = 28/57 (48%), Gaps = 10/57 (17%)
Frame = +2
Query: 186 KRPRPAAAGSPP----RRGSPPLHAPPRRY-----PGPPFDR-SPERPMNGYRSPPP 232
K P P PP + PP+++PP Y P PP+ SP P+ Y+SPPP
Sbjct: 227 KSPPPPVYSPPPVYKYKSPPPPVYSPPPPYKYKSPPPPPYKYPSPPPPVYKYKSPPP 397
Score = 34.3 bits (77), Expect = 0.081
Identities = 26/91 (28%), Positives = 36/91 (38%), Gaps = 15/91 (16%)
Frame = +2
Query: 188 PRPAAAGSPPRRGSPPLHAPPRRY---------------PGPPFDRSPERPMNGYRSPPP 232
P+P SPP PP+H+PP Y P PP P P+ Y+SPPP
Sbjct: 128 PKPYYYHSPP----PPVHSPPPPYHYSSPPPPPVYKYKSPPPPVYSPP--PVYKYKSPPP 289
Query: 233 RLMGRDGLRDYGPAAAALPPLRHEGRFPDPH 263
P + PP +++ P P+
Sbjct: 290 ------------PVYSPPPPYKYKSPPPPPY 346
Score = 30.8 bits (68), Expect = 0.90
Identities = 20/67 (29%), Positives = 29/67 (42%)
Frame = +2
Query: 199 RGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDGLRDYGPAAAALPPLRHEGR 258
+ PP P +YP PP P+ Y+SPPP + Y P PP ++ +
Sbjct: 413 KSPPPPPKKPYKYPSPP------PPVYKYKSPPPPV--------YSP-----PPPVYKYK 535
Query: 259 FPDPHLH 265
P P +H
Sbjct: 536 SPPPPVH 556
Score = 28.1 bits (61), Expect = 5.8
Identities = 16/43 (37%), Positives = 20/43 (46%)
Frame = +2
Query: 190 PAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPP 232
P P + SPP + P PP SP P+ Y+SPPP
Sbjct: 422 PPPPKKPYKYPSPPPPVYKYKSPPPPV-YSPPPPVYKYKSPPP 547
>TC81629 similar to GP|15081654|gb|AAK82482.1 AT4g26750/F10M23_90
{Arabidopsis thaliana}, partial (19%)
Length = 824
Score = 37.4 bits (85), Expect = 0.010
Identities = 32/107 (29%), Positives = 44/107 (40%), Gaps = 7/107 (6%)
Frame = +2
Query: 188 PRPAAAGSPPRRGSPPLHAPP--RRYPGPPF-DRSPERPMNGYRSPPPRLMGRDGLRDYG 244
P P+ PP S H PP + YP PP D P P Y+ PPP +DY
Sbjct: 17 PPPSQEYHPPPP-SQDYHPPPPSQDYPPPPSQDYHPPPPSQDYQPPPPS-------QDYH 172
Query: 245 PAAAALPPLRHEGRFPDPHLHRERMDYMDDAYRGRG----KFDRPPP 287
PP R + + +P+ H++ Y D + G + PPP
Sbjct: 173 QP----PPARSDSSYSEPYNHQQ---YSPDQSQNLGPNYPSHETPPP 292
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.143 0.479
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,602,780
Number of Sequences: 36976
Number of extensions: 343060
Number of successful extensions: 6124
Number of sequences better than 10.0: 325
Number of HSP's better than 10.0 without gapping: 3784
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5154
length of query: 379
length of database: 9,014,727
effective HSP length: 98
effective length of query: 281
effective length of database: 5,391,079
effective search space: 1514893199
effective search space used: 1514893199
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC122722.5


