

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122722.3 - phase: 0
(1085 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
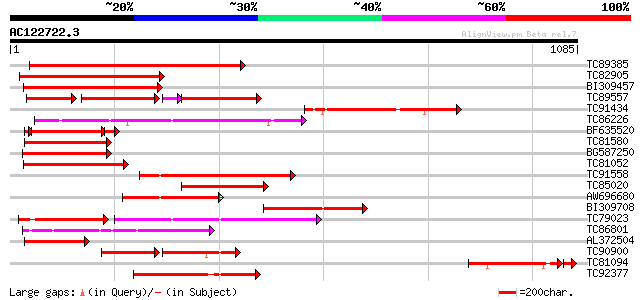
Score E
Sequences producing significant alignments: (bits) Value
TC89385 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resist... 612 e-175
TC82905 weakly similar to GP|3947733|emb|CAA08797.1 NL25 {Solanu... 556 e-158
BI309457 weakly similar to GP|15787901|gb resistance gene analog... 424 e-118
TC89557 weakly similar to GP|10178211|dbj|BAB11635. TMV resistan... 160 e-106
TC91434 weakly similar to PIR|T06144|T06144 disease resistance p... 375 e-104
TC86226 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 313 2e-85
BF635520 weakly similar to GP|3947735|emb| NL27 {Solanum tuberos... 247 5e-74
TC81580 weakly similar to GP|7341113|gb|AAF61210.1| unknown {Gly... 275 9e-74
BG587250 weakly similar to GP|3947735|emb| NL27 {Solanum tuberos... 270 2e-72
TC81052 weakly similar to GP|10178211|dbj|BAB11635. TMV resistan... 259 3e-69
TC91558 weakly similar to GP|19774201|gb|AAL99077.1 NBS-LRR-Toll... 254 1e-67
TC85020 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resist... 253 2e-67
AW696680 similar to GP|19774149|gb NBS-LRR-Toll resistance gene ... 235 8e-62
BI309708 weakly similar to PIR|D96753|D96 Similar to disease res... 234 1e-61
TC79023 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 223 3e-58
TC86801 similar to PIR|A54810|A54810 TMV resistance protein N - ... 215 6e-56
AL372504 weakly similar to GP|7341113|gb| unknown {Glycine max},... 213 4e-55
TC90900 weakly similar to PIR|D96753|D96753 Similar to disease r... 120 1e-52
TC81094 179 1e-51
TC92377 similar to GP|19774149|gb|AAL99051.1 NBS-LRR-Toll resist... 196 4e-50
>TC89385 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resistance gene
analog protein {Medicago ruthenica}, partial (98%)
Length = 1285
Score = 612 bits (1578), Expect = e-175
Identities = 307/414 (74%), Positives = 345/414 (83%)
Frame = +3
Query: 38 ALVTSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQKGECIGPELFR 97
ALV ++N YDVFVTFRGEDTRNNFT+FLF ALE KGI FRD NL KGE IGPEL R
Sbjct: 39 ALVPLPRRNCYDVFVTFRGEDTRNNFTNFLFAALERKGIYAFRDDTNLPKGESIGPELLR 218
Query: 98 AIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPSEVRKQSGIYS 157
IE SQV+VA+ S+NYASSTWCLQELEKICECIKGSGK+VLP+FY VDPSEV+KQSGIY
Sbjct: 219 TIEGSQVFVAVLSRNYASSTWCLQELEKICECIKGSGKYVLPIFYGVDPSEVKKQSGIYW 398
Query: 158 EAFVKHEQRFQQDSMKVSRWREALEQVGSISGWDLRDEPLAREIKEIVQKIINILECKYS 217
+ F KHEQRF+QD KVSRWREAL QVGSI+GWDLRD+ + E+++IVQ I+NIL+CK S
Sbjct: 399 DDFAKHEQRFKQDPHKVSRWREALNQVGSIAGWDLRDKQQSVEVEKIVQTILNILKCKSS 578
Query: 218 CVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLYGQISHQFSASC 277
VSKDLVGI+S +AL++ LLLNSVDGVR IGI GMGG GKTTLA LYGQI H+F ASC
Sbjct: 579 FVSKDLVGINSRTEALKHQLLLNSVDGVRVIGIWGMGGKGKTTLAMNLYGQICHRFDASC 758
Query: 278 FIDDVTKIYGLHDDPLDVQKQILFQTLGIEHQQICNRYHATTLIQRKLCHERTLMILDNV 337
FIDDV+KI+ LHD P+D QKQIL QTLGIEH QICN Y AT LI+ +L E+TL+ILDNV
Sbjct: 759 FIDDVSKIFRLHDGPIDAQKQILHQTLGIEHHQICNHYSATDLIRHRLSREKTLLILDNV 938
Query: 338 DQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVYKVSLLDWNEAHMLFCRKAF 397
DQVEQLE+I VHREWLG GSRI+IISRDEH+LK Y VDV YKV LLDW E+H LF +KAF
Sbjct: 939 DQVEQLERIGVHREWLGAGSRIVIISRDEHILKEYKVDVDYKVPLLDWTESHKLFRQKAF 1118
Query: 398 KDEKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFGRNVTEWKSALTRLRQSP 451
K EKIIM NYQNL +IL+YA GLPL I VLGSFL GRNVTEWKSAL RLRQ P
Sbjct: 1119KLEKIIMKNYQNLAYEILNYANGLPLTITVLGSFLSGRNVTEWKSALARLRQRP 1280
>TC82905 weakly similar to GP|3947733|emb|CAA08797.1 NL25 {Solanum
tuberosum}, partial (22%)
Length = 832
Score = 556 bits (1433), Expect = e-158
Identities = 277/277 (100%), Positives = 277/277 (100%)
Frame = +1
Query: 19 IQSSKHLYIQMASSSKSSSALVTSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMV 78
IQSSKHLYIQMASSSKSSSALVTSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMV
Sbjct: 1 IQSSKHLYIQMASSSKSSSALVTSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMV 180
Query: 79 FRDVINLQKGECIGPELFRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVL 138
FRDVINLQKGECIGPELFRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVL
Sbjct: 181 FRDVINLQKGECIGPELFRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVL 360
Query: 139 PVFYDVDPSEVRKQSGIYSEAFVKHEQRFQQDSMKVSRWREALEQVGSISGWDLRDEPLA 198
PVFYDVDPSEVRKQSGIYSEAFVKHEQRFQQDSMKVSRWREALEQVGSISGWDLRDEPLA
Sbjct: 361 PVFYDVDPSEVRKQSGIYSEAFVKHEQRFQQDSMKVSRWREALEQVGSISGWDLRDEPLA 540
Query: 199 REIKEIVQKIINILECKYSCVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGK 258
REIKEIVQKIINILECKYSCVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGK
Sbjct: 541 REIKEIVQKIINILECKYSCVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGK 720
Query: 259 TTLATTLYGQISHQFSASCFIDDVTKIYGLHDDPLDV 295
TTLATTLYGQISHQFSASCFIDDVTKIYGLHDDPLDV
Sbjct: 721 TTLATTLYGQISHQFSASCFIDDVTKIYGLHDDPLDV 831
>BI309457 weakly similar to GP|15787901|gb resistance gene analog NBS7
{Helianthus annuus}, partial (45%)
Length = 806
Score = 424 bits (1089), Expect = e-118
Identities = 211/266 (79%), Positives = 234/266 (87%)
Frame = +3
Query: 27 IQMASSSKSSSALVTSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQ 86
IQMAS+S SSS L TSS++N+YDVFVTFRGEDTRNNFTDFLFDAL+TKGI+VF D NL
Sbjct: 9 IQMASTSNSSSVLGTSSRRNYYDVFVTFRGEDTRNNFTDFLFDALQTKGIIVFSDDTNLP 188
Query: 87 KGECIGPELFRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDP 146
KGE IGPEL RAIE SQV+VA+FS NYASSTWCLQELEKICEC+KGSGKHVLPVFYDVDP
Sbjct: 189 KGESIGPELLRAIEGSQVFVAVFSINYASSTWCLQELEKICECVKGSGKHVLPVFYDVDP 368
Query: 147 SEVRKQSGIYSEAFVKHEQRFQQDSMKVSRWREALEQVGSISGWDLRDEPLAREIKEIVQ 206
S+VRKQSGIY EAF+KHEQRFQQ+ KVS+WR+AL+QVGSISGWDLRD+P A EIK+IVQ
Sbjct: 369 SDVRKQSGIYGEAFIKHEQRFQQEFQKVSKWRDALKQVGSISGWDLRDKPQAGEIKKIVQ 548
Query: 207 KIINILECKYSCVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLY 266
I+NIL+ K SC SKDLVGIDS + LQNHLLL+SVD VRAIGICGMGGIGKTTLA LY
Sbjct: 549 TILNILKYKSSCFSKDLVGIDSRLDGLQNHLLLDSVDSVRAIGICGMGGIGKTTLAMALY 728
Query: 267 GQISHQFSASCFIDDVTKIYGLHDDP 292
QISH+FSAS FIDDV + Y LHD P
Sbjct: 729 DQISHRFSASWFIDDVKQNY*LHDGP 806
>TC89557 weakly similar to GP|10178211|dbj|BAB11635. TMV resistance protein N
{Arabidopsis thaliana}, partial (8%)
Length = 1567
Score = 160 bits (406), Expect(4) = e-106
Identities = 75/153 (49%), Positives = 112/153 (73%)
Frame = +3
Query: 329 RTLMILDNVDQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVYKVSLLDWNEA 388
+ L++LDNV+Q+EQLEK+ + ++L P S+IIII+RD+H+L+AYG D V++ L++ +A
Sbjct: 1110 KLLIVLDNVEQLEQLEKLDIEPKFLHPRSKIIIITRDKHILQAYGADEVFEAKLMNDEDA 1289
Query: 389 HMLFCRKAFKDEKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFGRNVTEWKSALTRLR 448
H L CRKAFK + S + L+ ++L YA+ LPLA+KVLGSFLF RN EW S L +
Sbjct: 1290 HKLLCRKAFKSD-YPSSGFAELIPKVLVYAQRLPLAVKVLGSFLFSRNANEWSSTLDKFE 1466
Query: 449 QSPVKDVMDVLQLSFDGLNETEKDIFLHIACFF 481
++P +M LQ+S++GL + EK++FLH+A FF
Sbjct: 1467 KNPPNKIMKALQVSYEGLEKDEKEVFLHVAXFF 1565
Score = 156 bits (395), Expect(4) = e-106
Identities = 80/151 (52%), Positives = 106/151 (69%), Gaps = 1/151 (0%)
Frame = +1
Query: 137 VLPVFYDVDPSEVRKQSGIYSEAFVKHEQRFQQDSMKVSRWREALEQVGSISGWDLRDEP 196
V PVFYDVDPS V+KQ+G+Y AFV H + F+ +S KV+RWR + +G +GWD+R EP
Sbjct: 532 VFPVFYDVDPSHVKKQNGVYENAFVLHTEAFKHNSGKVARWRTTMTYLGGTAGWDVRVEP 711
Query: 197 LAREIKEIVQKIINILECKYSCVSKDLVGIDSPIQALQNHLLLNSV-DGVRAIGICGMGG 255
I++IV+ +I L +S + DL+GI I+AL+N L L+S DG R +GI GM G
Sbjct: 712 EFEMIEKIVEAVIKKLGRTFSGSTDDLIGIQPHIEALENLLKLSSKDDGCRVLGIWGMDG 891
Query: 256 IGKTTLATTLYGQISHQFSASCFIDDVTKIY 286
IGKTTLAT LY +IS QF A CFI++V+KIY
Sbjct: 892 IGKTTLATVLYDKISFQFDACCFIENVSKIY 984
Score = 103 bits (256), Expect(4) = e-106
Identities = 49/97 (50%), Positives = 67/97 (68%)
Frame = +3
Query: 32 SSKSSSALVTSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQKGECI 91
++ SSS S++ YDVF++FRG DTRN+F D L+ L KGI F+D LQKGE I
Sbjct: 216 TASSSSTDSNSNQSYRYDVFISFRGSDTRNSFVDHLYAHLNRKGIFTFKDDKQLQKGEAI 395
Query: 92 GPELFRAIEISQVYVAIFSKNYASSTWCLQELEKICE 128
P+L +AI+ S++ + +FSK+YASSTWCL E+ I E
Sbjct: 396 SPQLLQAIQQSRISIVVFSKDYASSTWCLDEMAAINE 506
Score = 27.7 bits (60), Expect(4) = e-106
Identities = 12/39 (30%), Positives = 23/39 (58%)
Frame = +2
Query: 292 PLDVQKQILFQTLGIEHQQICNRYHATTLIQRKLCHERT 330
P+ VQKQI+ QT+ ++ C+ + ++ +LC +T
Sbjct: 998 PVAVQKQIICQTIEEKNIDTCSARKISQTMRNRLCKTKT 1114
>TC91434 weakly similar to PIR|T06144|T06144 disease resistance protein
homolog F24J7.70 - Arabidopsis thaliana, partial (4%)
Length = 940
Score = 375 bits (963), Expect = e-104
Identities = 209/319 (65%), Positives = 233/319 (72%), Gaps = 19/319 (5%)
Frame = +1
Query: 565 HVEAIVLYYKEDEEADFEHLSKMSNLRLLFIA--------NYISTMLGFPSCLSNKLRFV 616
HVEA+V + D+ +F LS MSNLRLL I NY ML P LSNKLR+V
Sbjct: 1 HVEAVVFFGGIDKNVEF--LSTMSNLRLLIIRHDEYYMINNYELVMLK-PYSLSNKLRYV 171
Query: 617 HWFRYPSKYLPSNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDLRHSRNLEKIIDFG 676
W YP KYLPS+FHP ELVELIL S IKQLWKNKK+LPNLR LDL S+ LEKI DFG
Sbjct: 172 QWTGYPFKYLPSSFHPAELVELILVRSCIKQLWKNKKHLPNLRRLDLSDSKKLEKIEDFG 351
Query: 677 EFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMCG 736
+FPNLE L+LE CI LVELDPSIGLLRKLVYLNL+ C +LVSIPNNIFGLSSL+YLNM G
Sbjct: 352 QFPNLEWLNLERCIKLVELDPSIGLLRKLVYLNLERCYNLVSIPNNIFGLSSLKYLNMSG 531
Query: 737 CSKVFNNPRRLMKSGISSEKKQQHDIRESASH---HLPGLKWIILAHDSSHMLPSLHS-- 791
CSK LMK GISSEKK +HDIRES SH K I +++S P H+
Sbjct: 532 CSK-------LMKPGISSEKKNKHDIRESTSHCRSTSSVFKLFIFPNNASFSAPVTHTYK 690
Query: 792 ------LCCLRKVDISFCYLSHVPDAIECLHWLERLNLAGNDFVTLPSLRKLSKLVYLNL 845
L CLR +DISFC+LSHVPDAIECLH LERLNL GN+FVTLPS+RKLS+LVYLNL
Sbjct: 691 LPCFRILYCLRNIDISFCHLSHVPDAIECLHRLERLNLGGNNFVTLPSMRKLSRLVYLNL 870
Query: 846 EHCKLLESLPQLPFPTNTG 864
EHCKLLESLPQLPFP+ G
Sbjct: 871 EHCKLLESLPQLPFPSTIG 927
>TC86226 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (24%)
Length = 1769
Score = 313 bits (803), Expect = 2e-85
Identities = 191/536 (35%), Positives = 313/536 (57%), Gaps = 16/536 (2%)
Frame = +3
Query: 48 YDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDV--INLQKGECIGPELFRAIEISQVY 105
YDVF+ +G DTR FT L AL KGI F D +LQ+ + + P + IE S++
Sbjct: 90 YDVFLICKGTDTRYGFTGNLLKALIDKGIRTFHDDDDSDLQRRDKVTPII---IEESRIL 260
Query: 106 VAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPSEVRKQSGIYSEAFVKHEQ 165
+ IFS NYASS+ CL L I C K G VLPVF+ V+P++VR +G Y +A +HE
Sbjct: 261 IPIFSANYASSSSCLDTLVHIIHCYKTKGCLVLPVFFGVEPTDVRHHTGRYGKALAEHEN 440
Query: 166 RFQQDSMKVSR---WREALEQVGSISGWDLRDEPLAREIKEIVQKIINILECKYSCVSKD 222
RFQ D+ + R W+ AL ++ + D+ E E++ KI+ + K S S
Sbjct: 441 RFQNDTKNMERLQQWKVALSLAANLPSY--HDDSHGYEY-ELIGKIVKYISNKISRQSLH 611
Query: 223 L----VGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLYGQISHQFSASCF 278
+ VG+ S +Q +++ L DGV +GI G+GG GK+TLA +Y ++ QF CF
Sbjct: 612 VATYPVGLQSRVQQVKSLLDEGPDDGVHMVGIYGIGGSGKSTLARAIYNFVADQFEGLCF 791
Query: 279 IDDVTKIYGLHDDPLDVQKQILFQTLGIEHQQICNRYHATTLIQRKLCHERTLMILDNVD 338
++ V + + Q+ +L +TL ++ + + + ++I+ +LC ++ L+ILD+VD
Sbjct: 792 LEQVRE-NSASNSLKRFQEMLLSKTLQLKIK-LADVSEGISIIKERLCRKKILLILDDVD 965
Query: 339 QVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVYKVSLLDWNEAHMLFCRKAFK 398
++QL +A +W GPGSR+II +RD+H+L + ++ Y V L+ EA L AFK
Sbjct: 966 NMKQLNALAGGVDWFGPGSRVIITTRDKHLLACHEIEKTYAVKGLNVTEALELLRWMAFK 1145
Query: 399 DEKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFGRNVTEWKSALTRLRQSPVKDVMDV 458
++K+ S+Y+ ++++++ YA GLP+ I+++GS LFG+N+ E K+ L + P K++ +
Sbjct: 1146NDKV-PSSYEKILNRVVAYASGLPVVIEIVGSNLFGKNIEECKNTLDWYEKIPNKEIQRI 1322
Query: 459 LQLSFDGLNETEKDIFLHIACFFNNDSEEDVKNIL-----NCCGFHADIGLRVLIDKSLV 513
L++S+D L E E+ +FL IAC F E VK IL +C H + VL++K L+
Sbjct: 1323LKVSYDSLEEEEQSVFLDIACCFKGCKWEKVKEILHAHYGHCINHHVE----VLVEKCLI 1490
Query: 514 S-ISY-SIINMHSLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDVMLENMEKHVE 567
Y S +++H+L+E +G+++V+ S EP K SRLW + +++V+ EN ++
Sbjct: 1491DHFEYDSHVSLHNLIENMGKELVRLESPFEPGKRSRLWFEKDIFEVLEENTVSKID 1658
>BF635520 weakly similar to GP|3947735|emb| NL27 {Solanum tuberosum}, partial
(8%)
Length = 577
Score = 247 bits (631), Expect(3) = 5e-74
Identities = 121/144 (84%), Positives = 129/144 (89%)
Frame = +1
Query: 42 SSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQKGECIGPELFRAIEI 101
SSKKN+YDVFVT+RGEDTRNN TDFLFDALE+KGIMVFRD INLQKGE IG EL RAIE
Sbjct: 70 SSKKNYYDVFVTYRGEDTRNNLTDFLFDALESKGIMVFRDDINLQKGEYIGSELLRAIER 249
Query: 102 SQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPSEVRKQSGIYSEAFV 161
S VYVA+FS+NYASSTWCLQELEKICECI+G KHVLP+FYDVDPSEVRKQSGIY EAFV
Sbjct: 250 SHVYVAVFSRNYASSTWCLQELEKICECIEGLEKHVLPIFYDVDPSEVRKQSGIYWEAFV 429
Query: 162 KHEQRFQQDSMKVSRWREALEQVG 185
KHEQRFQQD VSRWREAL + G
Sbjct: 430 KHEQRFQQDFQMVSRWREALNKWG 501
Score = 49.3 bits (116), Expect(3) = 5e-74
Identities = 23/29 (79%), Positives = 26/29 (89%)
Frame = +3
Query: 182 EQVGSISGWDLRDEPLAREIKEIVQKIIN 210
+QVGSISGWDLRD+P A IK+IVQKIIN
Sbjct: 489 KQVGSISGWDLRDKPQAAXIKKIVQKIIN 575
Score = 22.3 bits (46), Expect(3) = 5e-74
Identities = 12/17 (70%), Positives = 13/17 (75%)
Frame = +3
Query: 29 MASSSKSSSALVTSSKK 45
MASSS SS ALVT K+
Sbjct: 30 MASSSNSSLALVTFFKE 80
>TC81580 weakly similar to GP|7341113|gb|AAF61210.1| unknown {Glycine max},
partial (84%)
Length = 827
Score = 275 bits (702), Expect = 9e-74
Identities = 135/168 (80%), Positives = 148/168 (87%)
Frame = +2
Query: 28 QMASSSKSSSALVTSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQK 87
QMASS+ SS ALVTSS+ N+YDVF TFRGEDTRNNFTDFLFDALETKGI FRD NLQK
Sbjct: 53 QMASSNNSSLALVTSSRNNYYDVFGTFRGEDTRNNFTDFLFDALETKGIFAFRDDTNLQK 232
Query: 88 GECIGPELFRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPS 147
GE I PEL RAIE S+V+VA+FS+NYASSTWCLQELEKIC+C++ S KH+LPVFYDVDPS
Sbjct: 233 GESIEPELLRAIEGSRVFVAVFSRNYASSTWCLQELEKICKCVQRSRKHILPVFYDVDPS 412
Query: 148 EVRKQSGIYSEAFVKHEQRFQQDSMKVSRWREALEQVGSISGWDLRDE 195
VRKQSGIY EAFVKHEQRFQQD VSRWREAL+ VGSISGWDLRD+
Sbjct: 413 VVRKQSGIYCEAFVKHEQRFQQDFEMVSRWREALKHVGSISGWDLRDK 556
>BG587250 weakly similar to GP|3947735|emb| NL27 {Solanum tuberosum}, partial
(9%)
Length = 655
Score = 270 bits (691), Expect = 2e-72
Identities = 133/171 (77%), Positives = 147/171 (85%)
Frame = +1
Query: 25 LYIQMASSSKSSSALVTSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVIN 84
LYIQMASSS SS+ALV ++N YDVFVTFRGEDTRNNFT+FLF ALE KGI FRD N
Sbjct: 1 LYIQMASSSNSSTALVPLPRRNCYDVFVTFRGEDTRNNFTNFLFAALERKGIYAFRDDTN 180
Query: 85 LQKGECIGPELFRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDV 144
L KGE IGPEL R IE SQV+VA+ S+NYASSTWCLQELEKICECIKGSGK+VLP+FY V
Sbjct: 181 LPKGESIGPELLRTIEGSQVFVAVLSRNYASSTWCLQELEKICECIKGSGKYVLPIFYGV 360
Query: 145 DPSEVRKQSGIYSEAFVKHEQRFQQDSMKVSRWREALEQVGSISGWDLRDE 195
DPSEV+KQSGIY + F KHEQRF+QD KVSRWREAL QVGSI+GWDLRD+
Sbjct: 361 DPSEVKKQSGIYWDDFAKHEQRFKQDPHKVSRWREALNQVGSIAGWDLRDK 513
>TC81052 weakly similar to GP|10178211|dbj|BAB11635. TMV resistance protein
N {Arabidopsis thaliana}, partial (8%)
Length = 656
Score = 259 bits (663), Expect = 3e-69
Identities = 133/204 (65%), Positives = 159/204 (77%), Gaps = 2/204 (0%)
Frame = +2
Query: 26 YIQMASSSKSSSALVTS--SKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVI 83
+IQMASSS SSS+ + + +KN+YDVFVTFRGEDTR NF D LF AL+ KGI FRD
Sbjct: 38 HIQMASSSNSSSSALMALPRRKNYYDVFVTFRGEDTRFNFIDHLFAALQRKGIFAFRDDA 217
Query: 84 NLQKGECIGPELFRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYD 143
NLQKGE I PEL RAIE SQV++A+ SKNY+SSTWCL+EL I +C + SG+ VLPVFYD
Sbjct: 218 NLQKGESIPPELIRAIEGSQVFIAVLSKNYSSSTWCLRELVHILDCSQVSGRRVLPVFYD 397
Query: 144 VDPSEVRKQSGIYSEAFVKHEQRFQQDSMKVSRWREALEQVGSISGWDLRDEPLAREIKE 203
VDPSEVR Q GIY EAF KHEQ FQ DS V WREAL QVG+ISGWDLRD+P EIK+
Sbjct: 398 VDPSEVRHQKGIYGEAFSKHEQTFQHDSHVVQSWREALTQVGNISGWDLRDKPQYAEIKK 577
Query: 204 IVQKIINILECKYSCVSKDLVGID 227
IV++I+NIL +S + K+LVG++
Sbjct: 578 IVEEILNILGHNFSSLPKELVGMN 649
>TC91558 weakly similar to GP|19774201|gb|AAL99077.1 NBS-LRR-Toll resistance
gene analog protein {Medicago edgeworthii}, partial
(60%)
Length = 942
Score = 254 bits (649), Expect = 1e-67
Identities = 134/302 (44%), Positives = 200/302 (65%), Gaps = 2/302 (0%)
Frame = +1
Query: 248 IGICGMGGIGKTTLATTLYGQISHQ--FSASCFIDDVTKIYGLHDDPLDVQKQILFQTLG 305
+GI M GIGKTTLA LY ISHQ F A CF++DV+KIY + VQK+IL QT+
Sbjct: 1 VGIWRMDGIGKTTLANVLYDTISHQYQFDACCFVEDVSKIYR-DGGAIAVQKRILDQTIK 177
Query: 306 IEHQQICNRYHATTLIQRKLCHERTLMILDNVDQVEQLEKIAVHREWLGPGSRIIIISRD 365
++ + + + +I +L + L++LDNVDQ QL+++ ++ L GSRIII +RD
Sbjct: 178 EKNLEGYSPSEISGIISNRLYKLKLLLVLDNVDQSVQLQELHINPISLCAGSRIIITTRD 357
Query: 366 EHVLKAYGVDVVYKVSLLDWNEAHMLFCRKAFKDEKIIMSNYQNLVDQILHYAKGLPLAI 425
+H+L Y D+VY+ LL+ N+AH L CRKAFK + S+Y+ L+ ++L YA+GLPLAI
Sbjct: 358 KHILIEYEADIVYEAELLNDNDAHELLCRKAFKSD-YSSSDYEELIPEVLKYAQGLPLAI 534
Query: 426 KVLGSFLFGRNVTEWKSALTRLRQSPVKDVMDVLQLSFDGLNETEKDIFLHIACFFNNDS 485
+V+GSFL+ R +W++AL + +P +M VL+ SF+GL EK+IFLH+ACFF+ +
Sbjct: 535 RVMGSFLYKRKTAQWRAALEGWQNNPDSGIMKVLRSSFEGLELREKEIFLHVACFFDGER 714
Query: 486 EEDVKNILNCCGFHADIGLRVLIDKSLVSISYSIINMHSLLEELGRKIVQNSSSKEPRKW 545
E+ V+ IL+ CG +IG+ ++KSL++I +MH +L+ELG V+ + EPR W
Sbjct: 715 EDYVRRILHACGLQPNIGIPTSVEKSLITIRNQENHMHEMLQELGETNVRETHPDEPRLW 894
Query: 546 SR 547
S+
Sbjct: 895 SK 900
>TC85020 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resistance gene
analog protein {Medicago ruthenica}, partial (55%)
Length = 500
Score = 253 bits (647), Expect = 2e-67
Identities = 125/166 (75%), Positives = 144/166 (86%)
Frame = +1
Query: 329 RTLMILDNVDQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVYKVSLLDWNEA 388
+ L+I DNVDQVEQLEKIAVHREWLG GSRI+IISRDEH+LK YGVDVVYKV L++ ++
Sbjct: 1 KALLIFDNVDQVEQLEKIAVHREWLGAGSRIVIISRDEHILKEYGVDVVYKVPLMNSTDS 180
Query: 389 HMLFCRKAFKDEKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFGRNVTEWKSALTRLR 448
+ LFCRKAFK EKIIMS+YQNL ++IL YAKGLPLAIKVLGSFLFG +V EWKSAL RLR
Sbjct: 181 YELFCRKAFKVEKIIMSDYQNLANEILDYAKGLPLAIKVLGSFLFGHSVAEWKSALARLR 360
Query: 449 QSPVKDVMDVLQLSFDGLNETEKDIFLHIACFFNNDSEEDVKNILN 494
+SP DVMDVL LSFDGL+E EK+IFL IACFFN+ E+ VKN+LN
Sbjct: 361 ESPHNDVMDVLHLSFDGLDEKEKEIFLDIACFFNSQPEKYVKNVLN 498
>AW696680 similar to GP|19774149|gb NBS-LRR-Toll resistance gene analog
protein {Medicago sativa}, partial (96%)
Length = 641
Score = 235 bits (599), Expect = 8e-62
Identities = 121/193 (62%), Positives = 149/193 (76%)
Frame = +2
Query: 216 YSCVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLYGQISHQFSA 275
YS + K+LVG++S I + N LLL+S+D VR +GICGMGGIGKTTLAT LYGQISHQF A
Sbjct: 5 YSSLPKELVGMNSHIDKVANLLLLDSIDDVRVVGICGMGGIGKTTLATALYGQISHQFDA 184
Query: 276 SCFIDDVTKIYGLHDDPLDVQKQILFQTLGIEHQQICNRYHATTLIQRKLCHERTLMILD 335
CFIDD++KIY HD + QKQIL QTLG+E Q+CN YH T L++R+L R L+I+D
Sbjct: 185 RCFIDDLSKIYR-HDGQVGAQKQILHQTLGVEPFQLCNLYHTTDLMRRRLRRLRVLIIVD 361
Query: 336 NVDQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVYKVSLLDWNEAHMLFCRK 395
NVD+V QL+K+ V+REWLG GSRIIIIS DEH+LK YGVDVVY+V LL+W + LF K
Sbjct: 362 NVDKVGQLDKLGVNREWLGAGSRIIIISGDEHILKEYGVDVVYRVPLLNWTNSLQLFSLK 541
Query: 396 AFKDEKIIMSNYQ 408
AFK II S+Y+
Sbjct: 542 AFKLYHII-SDYE 577
>BI309708 weakly similar to PIR|D96753|D96 Similar to disease resistance
proteins [imported] - Arabidopsis thaliana, partial (4%)
Length = 621
Score = 234 bits (598), Expect = 1e-61
Identities = 126/203 (62%), Positives = 154/203 (75%), Gaps = 4/203 (1%)
Frame = +1
Query: 486 EEDVKNILNCCGFHADIGLRVLIDKSLVSISY-SIINMHSLLEELGRKIVQNSSSKEPRK 544
E+ VKN+LNCCGFHADIGLRVLIDKS++SIS + I +H LL+ELGRKIVQ S KE RK
Sbjct: 13 EKYVKNVLNCCGFHADIGLRVLIDKSVISISTENNIEIHRLLQELGRKIVQEKSIKESRK 192
Query: 545 WSRLWSTEQLYDVMLENMEKHVEAIVLYYKEDEEADF---EHLSKMSNLRLLFIANYIST 601
WSR+W +Q Y+VM ENMEK V AIV + E F E LSKM +LRLL + T
Sbjct: 193 WSRMWLHKQFYNVMSENMEKKVGAIVFVRDKKERKIFIMAETLSKMIHLRLLILKGV--T 366
Query: 602 MLGFPSCLSNKLRFVHWFRYPSKYLPSNFHPNELVELILTESNIKQLWKNKKYLPNLRTL 661
+ G + LS++LR+V W RYP KYLPS+F PN+LVELIL S++KQLWK+KKYLPNLRTL
Sbjct: 367 LTGNLNGLSDELRYVEWNRYPFKYLPSSFLPNQLVELILRYSSVKQLWKDKKYLPNLRTL 546
Query: 662 DLRHSRNLEKIIDFGEFPNLERL 684
DL HS++L K+ +FGE PNLER+
Sbjct: 547 DLSHSKSLRKMPNFGEVPNLERV 615
>TC79023 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (37%)
Length = 2072
Score = 223 bits (568), Expect = 3e-58
Identities = 135/404 (33%), Positives = 242/404 (59%), Gaps = 7/404 (1%)
Frame = +3
Query: 201 IKEIVQKIINILECKYSCVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTT 260
I +IV+ I N + + V+K VG+ S +Q ++ HL S D V +G+ G GGIGK+T
Sbjct: 705 IGKIVEDISNRISREPLDVAKYPVGLQSRVQHVKGHLDEKSDDEVHMVGLYGTGGIGKST 884
Query: 261 LATTLYGQISHQFSASCFIDDVTKIYGLHDDPLDVQKQILFQTLGIEHQQICNRYHATTL 320
LA +Y I+ QF CF+++V ++ D+ +Q+++L +T+ ++ + + +
Sbjct: 885 LAKAIYNFIADQFEVLCFLENV-RVNSTSDNLKHLQEKLLLKTVRLDIK-LGGVSQGIPI 1058
Query: 321 IQRKLCHERTLMILDNVDQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVYKV 380
I+++LC ++ L+ILD+VD+++QLE +A +W GPGSR+II +R++H+LK +G++ + V
Sbjct: 1059 IKQRLCRKKILLILDDVDKLDQLEALAGGLDWFGPGSRVIITTRNKHLLKIHGIESTHAV 1238
Query: 381 SLLDWNEAHMLFCRKAFKDEKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFGRNVTEW 440
L+ EA L AFK+ + S++++++++ L YA GLPLAI ++GS L GR+V +
Sbjct: 1239 EGLNATEALELLRWMAFKEN--VPSSHEDILNRALTYASGLPLAIVIIGSNLVGRSVQDS 1412
Query: 441 KSALTRLRQSPVKDVMDVLQLSFDGLNETEKDIFLHIACFFNNDSEEDVKNILNCCGFHA 500
S L + P K++ +L++S+D L + E+ +FL IAC F +VK IL+ H
Sbjct: 1413 MSTLDGYEEIPNKEIQRILKVSYDSLEKEEQSVFLDIACCFKGCKWPEVKEILHAHYGHC 1592
Query: 501 DI-GLRVLIDKSLVS-ISY-SIINMHSLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDV 557
+ + VL +KSL+ + Y S + +H L+E++G+++V+ S EP + SRLW + V
Sbjct: 1593 IVHHVAVLAEKSLMDHLKYDSYVTLHDLIEDMGKEVVRQESPDEPGERSRLWFERDIVHV 1772
Query: 558 MLENM-EKHVEAIVLYY---KEDEEADFEHLSKMSNLRLLFIAN 597
+ +N + ++ I + + + D + + KM+NL+ N
Sbjct: 1773 LKKNTGTRKIKMINMKFPSMESDIDWNGNAFEKMTNLKTFITEN 1904
Score = 138 bits (347), Expect = 1e-32
Identities = 73/175 (41%), Positives = 109/175 (61%), Gaps = 3/175 (1%)
Frame = +1
Query: 18 HIQSSKHLYIQMASSSKSSSALVTSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIM 77
H Q+ H ++ + +++S S+ Y VF++FRG DTR+ FT L+ AL KGI
Sbjct: 25 HSQTPCHKFLSVLMATQSPSSFT-------YQVFLSFRGADTRHGFTGNLYKALTDKGIY 183
Query: 78 VFRDVINLQKGECIGPELFRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHV 137
F D +LQ+G+ I P L AIE S++++ +FS+NYASS++CL EL I C G V
Sbjct: 184 TFIDDNDLQRGDEITPSLKNAIEKSRIFIPVFSENYASSSFCLDELVHITHCYDTKGCLV 363
Query: 138 LPVFYDVDPSEVRKQSGIYSEAFVKHEQRFQQD---SMKVSRWREALEQVGSISG 189
LPVF VDP++VR +G Y EA H+++FQ D + ++ +W+EAL Q ++SG
Sbjct: 364 LPVFIGVDPTDVRHHTGRYGEALAVHKKKFQNDKDNTERLQQWKEALSQAANLSG 528
>TC86801 similar to PIR|A54810|A54810 TMV resistance protein N - tobacco
(Nicotiana glutinosa), partial (7%)
Length = 1268
Score = 215 bits (548), Expect = 6e-56
Identities = 139/376 (36%), Positives = 216/376 (56%), Gaps = 7/376 (1%)
Frame = +2
Query: 24 HLYIQMASSSKSSSALVTSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVI 83
H MA S SSS S+ + YDVF++FRGEDTR FT L AL + + D
Sbjct: 5 HTS*NMAMSLASSSH---SAAQKKYDVFISFRGEDTRAGFTSHLHAALSRTYLHTYIDY- 172
Query: 84 NLQKGECIGPELFRAIEISQVYVAIFSKNYASSTWCLQELEKICEC---IKGSGKHVLPV 140
++KG+ + PEL +AI+ S +++ +FS+NYASSTWCL EL ++ EC + V+PV
Sbjct: 173 RIEKGDEVWPELEKAIKQSTLFLVVFSENYASSTWCLNELVELMECRNKNEDDNIGVIPV 352
Query: 141 FYDVDPSEVRKQSGIYSEAFVKHEQRFQQDSMKVSRWREALEQVGSISGW---DLRDEPL 197
FY VDPS VRKQ+G Y A KH+Q Q D M + W+ AL Q ++SG+ R E
Sbjct: 353 FYHVDPSHVRKQTGSYGSALAKHKQENQDDKM-MQNWKNALFQAANLSGFHSSTYRTE-- 523
Query: 198 AREIKEIVQKIINILECKYSCVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIG 257
+ I++I + ++ L +Y + +D A+++ + +S V+ IG+ GMGG G
Sbjct: 524 SNMIEDITRALLGKLNHQYRDELTCNLILDENYWAVRSLIKFDSTT-VQIIGLWGMGGTG 700
Query: 258 KTTLATTLYGQISHQFSASCFIDDVTKIYGLHDDPLDVQKQILFQTLGIEHQQICNRYHA 317
KTTLA ++ + S ++ +CF++ VT++ H K +L + LG E +I
Sbjct: 701 KTTLAAAMFQRFSFKYEGNCFLERVTEVSKKHGINYTCNK-LLSKLLG-EDLRIDTPKVI 874
Query: 318 TTLIQRKLCHERTLMILDNVDQVEQLEK-IAVHREWLGPGSRIIIISRDEHVLKAYGVDV 376
+I+R+L H ++ ++LD+V E L+ I V WLGPGS +I+ +RD+HVL + G+D
Sbjct: 875 PAMIKRRLRHMKSFIVLDDVHNSELLQDLIGVRGGWLGPGSIVIVTTRDKHVLISGGIDE 1054
Query: 377 VYKVSLLDWNEAHMLF 392
+Y+V ++ + LF
Sbjct: 1055IYEVKKMNSQNSLQLF 1102
>AL372504 weakly similar to GP|7341113|gb| unknown {Glycine max}, partial
(70%)
Length = 478
Score = 213 bits (541), Expect = 4e-55
Identities = 102/125 (81%), Positives = 114/125 (90%)
Frame = +2
Query: 28 QMASSSKSSSALVTSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQK 87
QMA+SS SS ALVTSS++N+YDVFVTFRGEDTRNNFTD+LFDALETKGI FRD NL+K
Sbjct: 104 QMANSSNSSLALVTSSRRNYYDVFVTFRGEDTRNNFTDYLFDALETKGIYAFRDDTNLKK 283
Query: 88 GECIGPELFRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPS 147
GE IGPEL RAIE SQV+VA+FS+NYASSTWCLQELEKICEC++G KHVLPVFYD+DPS
Sbjct: 284 GEVIGPELLRAIEGSQVFVAVFSRNYASSTWCLQELEKICECVQGPEKHVLPVFYDIDPS 463
Query: 148 EVRKQ 152
EVRKQ
Sbjct: 464 EVRKQ 478
>TC90900 weakly similar to PIR|D96753|D96753 Similar to disease resistance
proteins [imported] - Arabidopsis thaliana, partial (7%)
Length = 826
Score = 120 bits (302), Expect(2) = 1e-52
Identities = 64/157 (40%), Positives = 104/157 (65%), Gaps = 8/157 (5%)
Frame = +3
Query: 293 LDVQKQILFQTLGIEHQQICNRYHATTLIQRKLCHERTLMILDNVDQVEQLEKIAVHREW 352
+ +QKQIL QT+ ++ + + + +++++LC+ + L++LDNVD +EQ+E++A++ E
Sbjct: 357 VSLQKQILRQTIDEKYLETYSPSEISGIVRKRLCNRKFLVVLDNVDLLEQVEELAINPEL 536
Query: 353 LGPGSRIIIISRDEHVLKAYGVD--------VVYKVSLLDWNEAHMLFCRKAFKDEKIIM 404
+G GSR+II +R+ H+L+ YG V Y+V LL+ N+A LF RKAFK +
Sbjct: 537 VGKGSRMIITTRNMHILRVYGEQLSLSHGTCVSYEVPLLNNNDARELFYRKAFKSQD-PA 713
Query: 405 SNYQNLVDQILHYAKGLPLAIKVLGSFLFGRNVTEWK 441
S NL ++L Y +GLPLAI+V+GSFL RN +W+
Sbjct: 714 SECLNLTPEVLKYVEGLPLAIRVVGSFLCTRNANQWR 824
Score = 105 bits (263), Expect(2) = 1e-52
Identities = 53/112 (47%), Positives = 79/112 (70%), Gaps = 1/112 (0%)
Frame = +2
Query: 176 RWREALEQVGSISGWDLRDEPLAREIKEIVQKIINILECKYSCVSKDLVGIDSPIQALQN 235
RW +A+ ++ + GWD+R++P REI+ IVQ++I L K+S + DL+ ++ L++
Sbjct: 5 RWTKAMGRLAELVGWDVRNKPEFREIENIVQEVIKTLGHKFSGFADDLIATQPRVEELES 184
Query: 236 HLLLNSVDG-VRAIGICGMGGIGKTTLATTLYGQISHQFSASCFIDDVTKIY 286
L L+S D +R +GI GM GIGKTTLA+ LY +IS QF ASCFI++V+KIY
Sbjct: 185 LLKLSSDDDELRVVGIWGMAGIGKTTLASVLYDRISSQFDASCFIENVSKIY 340
>TC81094
Length = 1448
Score = 179 bits (454), Expect(2) = 1e-51
Identities = 94/191 (49%), Positives = 118/191 (61%), Gaps = 10/191 (5%)
Frame = +2
Query: 878 GLLIFNCPKLGEREHCRSMTLLWMKQFIKANPRSSS----EIQIVNPGSEIPSWINNQRM 933
GL+IFNCPKLGERE C S+T WMKQFI+AN +S E+QIV PGSEIPSWINNQ M
Sbjct: 11 GLVIFNCPKLGERECCSSITFSWMKQFIQANQQSYGPYLYELQIVTPGSEIPSWINNQSM 190
Query: 934 GYSIAIDRSPIRHDNDNNIIGIVCCAAFTMAPYREIFYSSELMNLAFKRIDSNERLLKMR 993
G SI ID SP+ HDN NNIIG V CA F MAP + +++ + ++ K
Sbjct: 191 GGSILIDESPVIHDNKNNIIGFVFCAVFCMAPQDQTMIECLPLSV-YMKMGDERNCRKFP 367
Query: 994 VPVKLSLVTTKSSHLWIIYLPREYPG------YSCHEFGKIELKFFEVEGLEVESCGYRW 1047
V + L+ TKSSHLW++Y PREY C +G+ +V G++V+ CGYRW
Sbjct: 368 VIIDRDLIPTKSSHLWLVYFPREYYDVFGTIRIYCTRYGR------QVVGMDVKCCGYRW 529
Query: 1048 VCKQDIQEFNL 1058
VCKQ + L
Sbjct: 530 VCKQKLTRVQL 562
Score = 43.9 bits (102), Expect(2) = 1e-51
Identities = 18/25 (72%), Positives = 21/25 (84%)
Frame = +1
Query: 1060 MNHKNSLTPKCKILTIEDETQPEPQ 1084
+NH+ SL KCKIL IEDETQP+PQ
Sbjct: 571 LNHEKSLASKCKILAIEDETQPQPQ 645
>TC92377 similar to GP|19774149|gb|AAL99051.1 NBS-LRR-Toll resistance gene
analog protein {Medicago sativa}, partial (57%)
Length = 945
Score = 196 bits (498), Expect = 4e-50
Identities = 118/243 (48%), Positives = 151/243 (61%)
Frame = +1
Query: 237 LLLNSVDGVRAIGICGMGGIGKTTLATTLYGQISHQFSASCFIDDVTKIYGLHDDPLDVQ 296
LLL S D ++ +GI GMGGIGKTTLAT LY +ISHQF A CFIDD++K+Y ++ L ++
Sbjct: 232 LLLESTDIIQVVGIYGMGGIGKTTLATALYDRISHQFDACCFIDDISKVY-RNEGRLVLK 408
Query: 297 KQILFQTLGIEHQQICNRYHATTLIQRKLCHERTLMILDNVDQVEQLEKIAVHREWLGPG 356
+ + L + T LI+ KL H R L++LDNVDQVEQ EK+AV E LG G
Sbjct: 409 SKHYIKLLANSTFRYAIFTTKTNLIRLKLGHIRALLVLDNVDQVEQQEKLAVSSECLGAG 588
Query: 357 SRIIIISRDEHVLKAYGVDVVYKVSLLDWNEAHMLFCRKAFKDEKIIMSNYQNLVDQILH 416
SRI+I SRD H+LK Y VD + FC+KAF+ K I SNY+ L I
Sbjct: 589 SRIVITSRDVHILKEYEVDEIVN-----------*FCQKAFQGGK-IKSNYEELTCHI-K 729
Query: 417 YAKGLPLAIKVLGSFLFGRNVTEWKSALTRLRQSPVKDVMDVLQLSFDGLNETEKDIFLH 476
+G L GRN++EW SAL RLR++P KD+MDVL+LSF+GL E+E IFL
Sbjct: 730 LC*WPTTGN*SIGLILCGRNISEWTSALARLRENPNKDIMDVLRLSFEGLEESENVIFLD 909
Query: 477 IAC 479
I C
Sbjct: 910 IVC 918
Score = 40.4 bits (93), Expect = 0.004
Identities = 41/132 (31%), Positives = 58/132 (43%), Gaps = 16/132 (12%)
Frame = +3
Query: 201 IKEIVQKIINILECKYSCVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGI------CGMG 254
I++IV++IIN L ++S + KDLVGIDS I+ L+ V GI C
Sbjct: 123 IEKIVEEIINRLGYRFSSLPKDLVGIDSLIEELEK---------VFTFGIN*YYTGCRNL 275
Query: 255 GIGKTTLATTLYGQISHQFSAS-CFIDDVTKIYGLHD---------DPLDVQKQILFQTL 304
G+ + Y + S+ C + LH + QKQ L QTL
Sbjct: 276 WNGRNRKDDSSYCSL*QNLSSI*CML--------LHR*YKQSL*K*RSIGAQKQALHQTL 431
Query: 305 GIEHQQICNRYH 316
+H QICN Y+
Sbjct: 432 SKQHLQICNLYN 467
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.138 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 39,329,883
Number of Sequences: 36976
Number of extensions: 624901
Number of successful extensions: 4061
Number of sequences better than 10.0: 332
Number of HSP's better than 10.0 without gapping: 3707
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3866
length of query: 1085
length of database: 9,014,727
effective HSP length: 106
effective length of query: 979
effective length of database: 5,095,271
effective search space: 4988270309
effective search space used: 4988270309
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC122722.3


