

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122169.1 + phase: 0 /pseudo
(459 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
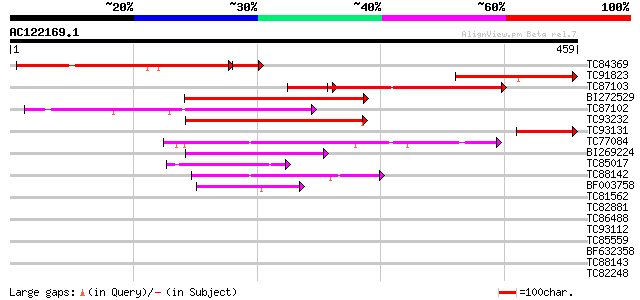
Score E
Sequences producing significant alignments: (bits) Value
TC84369 similar to GP|19310729|gb|AAL85095.1 unknown protein {Ar... 281 3e-84
TC91823 weakly similar to GP|19310729|gb|AAL85095.1 unknown prot... 187 1e-47
TC87103 similar to GP|15292735|gb|AAK92736.1 unknown protein {Ar... 137 3e-38
BI272529 weakly similar to GP|6478925|gb| hypothetical protein {... 149 3e-36
TC87102 similar to PIR|T02589|T02589 hypothetical protein T16B24... 147 6e-36
TC93232 weakly similar to GP|21739227|emb|CAD40982. OSJNBa0072F1... 140 1e-33
TC93131 similar to GP|19310729|gb|AAL85095.1 unknown protein {Ar... 77 1e-14
TC77084 similar to GP|15529155|gb|AAK97672.1 AT3g30390/T6J22_16 ... 60 2e-09
BI269224 weakly similar to GP|11862972|db hypothetical protein {... 51 1e-06
TC85017 weakly similar to GP|9758059|dbj|BAB08638.1 amino acid t... 49 3e-06
TC88142 weakly similar to GP|14587360|dbj|BAB61261. contains EST... 45 6e-05
BF003758 similar to GP|15216030|em amino acid permease AAP4 {Vic... 42 6e-04
TC81562 weakly similar to GP|6671946|gb|AAF23206.1| putative ami... 40 0.001
TC82881 similar to GP|15216026|emb|CAC51423. amino acid permease... 39 0.003
TC86488 similar to GP|9665144|gb|AAF97328.1| Unknown protein {Ar... 35 0.046
TC93112 weakly similar to GP|6671946|gb|AAF23206.1| putative ami... 35 0.079
TC85559 similar to PIR|T07012|T07012 acetyl-CoA carboxylase (EC ... 35 0.079
BF632358 weakly similar to GP|19352296|gb MULTIPLE BANDED ANTIGE... 33 0.18
TC88143 weakly similar to GP|20453045|gb|AAM19768.1 AT3g56200/F1... 33 0.23
TC82248 similar to GP|15216028|emb|CAC51424. amino acid permease... 33 0.23
>TC84369 similar to GP|19310729|gb|AAL85095.1 unknown protein {Arabidopsis
thaliana}, partial (8%)
Length = 614
Score = 281 bits (719), Expect(2) = 3e-84
Identities = 138/182 (75%), Positives = 157/182 (85%), Gaps = 6/182 (3%)
Frame = +3
Query: 6 SDKNKEKENDYFLDANEDETDIEAVNYDSDSSNNNDDDDEDDRIATHRPESFTSQQWPQS 65
SDKNKEKEN+YFLDANEDETDIEAVNYDSDSSNN D+DD+ +PESFTSQQWPQ+
Sbjct: 6 SDKNKEKENEYFLDANEDETDIEAVNYDSDSSNNRGSYDDDDQ----QPESFTSQQWPQT 173
Query: 66 YNEALDPLTIAAAPNIGSVLRAPSVIYASFAAGRSSKSYLELQDG---FLTGTQIQE--- 119
YNEA+DPLTIAAAPN+GS+LR PSV+Y+SF SSKSYLEL DG FL+G QIQE
Sbjct: 174 YNEAIDPLTIAAAPNLGSILRGPSVLYSSFVGSFSSKSYLELHDGKTSFLSGNQIQEGFP 353
Query: 120 STWWEKASIQKNIPEELPIGYGCTFTQTIFNGLNVLAGVGLLSAPDTVKQAGWASLLVIV 179
+TWWEKASIQ IPEELPIGYGCTFTQTIFNG+N++AGVGLLS PDTVKQAGW SL+V++
Sbjct: 354 TTWWEKASIQMQIPEELPIGYGCTFTQTIFNGINIMAGVGLLSTPDTVKQAGWVSLVVML 533
Query: 180 VF 181
+F
Sbjct: 534 IF 539
Score = 49.3 bits (116), Expect(2) = 3e-84
Identities = 22/25 (88%), Positives = 22/25 (88%)
Frame = +1
Query: 181 FAVVCFYTAELMRHCFQSREGIISY 205
FAVVC YTA LM HCFQSREGIISY
Sbjct: 538 FAVVCXYTAXLMXHCFQSREGIISY 612
>TC91823 weakly similar to GP|19310729|gb|AAL85095.1 unknown protein
{Arabidopsis thaliana}, partial (20%)
Length = 926
Score = 187 bits (474), Expect = 1e-47
Identities = 95/104 (91%), Positives = 95/104 (91%), Gaps = 6/104 (5%)
Frame = +3
Query: 362 LICFVLPVFLYGSVGAAGFLMFGERTSSQITLDLPRDAFASKVSLWTIT------HML** 415
L FVLPVFLYGSVGAAGFLMFGERTSSQITLDLPRDAFASKVSLWTI HML**
Sbjct: 39 LYSFVLPVFLYGSVGAAGFLMFGERTSSQITLDLPRDAFASKVSLWTIAIIPLTKHML** 218
Query: 416 *THWQEVWRNCCLIASHALIGASFFSEQHWLYLQFVLLF*FHFS 459
*THWQEVWRNCCLIASHALIGASFFSEQHWLYLQFVLLF*FHFS
Sbjct: 219 *THWQEVWRNCCLIASHALIGASFFSEQHWLYLQFVLLF*FHFS 350
>TC87103 similar to GP|15292735|gb|AAK92736.1 unknown protein {Arabidopsis
thaliana}, partial (51%)
Length = 1621
Score = 137 bits (344), Expect(2) = 3e-38
Identities = 66/145 (45%), Positives = 94/145 (64%)
Frame = +3
Query: 258 LNLDGKHLFAILAALIILPTVWLKDLRFVSYLSAGGVVGTALVGACVYAVGTRKDVGFHH 317
LN + + LFA++AAL +LPTVWL+DL +SY+SAGGV+ + LV C+ +G +DVGF
Sbjct: 507 LNXNPQTLFAVVAALAVLPTVWLRDLSVLSYISAGGVIASVLVVLCLLWIGI-EDVGFQR 683
Query: 318 TAPLVNWSGVPFAFGIYGFCFAGHSVFPNIYQSMANKKDFTKAMLICFVLPVFLYGSVGA 377
+ +N +P A G+YG+C++GH+VFPNIY SMA F ++ CF + LY
Sbjct: 684 SGTTLNLGTLPVAIGLYGYCYSGHAVFPNIYTSMAKPNQFPAVLVACFGVCTLLYAGGAV 863
Query: 378 AGFLMFGERTSSQITLDLPRDAFAS 402
G+ GE T SQ TL+LP+D A+
Sbjct: 864 MGYKNVGEDTLSQFTLNLPQDLVAT 938
Score = 39.7 bits (91), Expect(2) = 3e-38
Identities = 15/40 (37%), Positives = 25/40 (62%)
Frame = +2
Query: 226 YTELYSYCVEFIIMEGDNLSGLFPGTSLHWGSLNLDGKHL 265
Y + C+E+II+EGDNL+ LFP L+ G + + ++
Sbjct: 410 YVVMQGCCIEYIILEGDNLASLFPNAYLNLGGIEXESTNI 529
>BI272529 weakly similar to GP|6478925|gb| hypothetical protein {Arabidopsis
thaliana}, partial (23%)
Length = 625
Score = 149 bits (375), Expect = 3e-36
Identities = 70/149 (46%), Positives = 102/149 (67%)
Frame = +2
Query: 142 CTFTQTIFNGLNVLAGVGLLSAPDTVKQAGWASLLVIVVFAVVCFYTAELMRHCFQSREG 201
C+F +++ NG N+L G+GL++ P VK+ G+ SL ++++FAV+C YT L+ C QS G
Sbjct: 131 CSFARSVINGTNLLCGIGLMTMPYAVKEGGFLSLALLLLFAVICCYTGILLMRCLQSYPG 310
Query: 202 IISYPDIGEAAFGKYGRVFISIVLYTELYSYCVEFIIMEGDNLSGLFPGTSLHWGSLNLD 261
+ +YPDIG+AAFG GR+ I+I+L EL+ VEF+ + DNLS LFP TS+ L
Sbjct: 311 LDTYPDIGQAAFGYAGRLGIAIILSLELFGASVEFLTLVSDNLSALFPNTSMIVAGTELS 490
Query: 262 GKHLFAILAALIILPTVWLKDLRFVSYLS 290
+F I AAL++LPT WLK+L +SY+S
Sbjct: 491 THQVFTIAAALLVLPTGWLKNLSLLSYIS 577
>TC87102 similar to PIR|T02589|T02589 hypothetical protein T16B24.23 -
Arabidopsis thaliana (fragment), partial (44%)
Length = 964
Score = 147 bits (372), Expect = 6e-36
Identities = 84/256 (32%), Positives = 136/256 (52%), Gaps = 20/256 (7%)
Frame = +2
Query: 13 ENDYFLDANE-DETDIEAVNYDSDSSNNNDDDDEDDRIATHRPESFTSQQWPQSYNEALD 71
EN + ++++E D+ D D N++D + + R +S + WPQSY +++D
Sbjct: 176 ENSFIIESDEEDDKDFNK----GDDGNDSDSSNYSNENPPQRKQSSYNPSWPQSYRQSID 343
Query: 72 PLTIAAAPNIG-----SVLRAPSVIYASFAAGRSSKSYL-ELQDGFLTGTQIQESTWWEK 125
+ +P+IG S+ R S ++ R + L + + T+ ++
Sbjct: 344 LYSSVPSPSIGFLGNSSLTRLSSSFLSTSLTRRHTPEVLPSVTKPLIQPTEEEKHQRRSS 523
Query: 126 ASI-------------QKNIPEELPIGYGCTFTQTIFNGLNVLAGVGLLSAPDTVKQAGW 172
++ + + E+P + C+F Q + NG+NVL GVG+LS P K+ GW
Sbjct: 524 HTLLPPLSRRSSLLKKESKVSHEVPSRH-CSFGQAVLNGINVLCGVGILSTPYAAKEGGW 700
Query: 173 ASLLVIVVFAVVCFYTAELMRHCFQSREGIISYPDIGEAAFGKYGRVFISIVLYTELYSY 232
L ++ +F ++ FYT L+R C S G+ +YPDIG+AAFG GR+ ISIVLY ELY
Sbjct: 701 LGLSILFIFGILSFYTGLLLRSCLDSEPGLETYPDIGQAAFGTAGRIAISIVLYVELYGC 880
Query: 233 CVEFIIMEGDNLSGLF 248
C+E+I +EGDNL+ LF
Sbjct: 881 CIEYINLEGDNLAFLF 928
>TC93232 weakly similar to GP|21739227|emb|CAD40982. OSJNBa0072F16.6 {Oryza
sativa}, partial (56%)
Length = 695
Score = 140 bits (352), Expect = 1e-33
Identities = 69/149 (46%), Positives = 92/149 (61%), Gaps = 2/149 (1%)
Frame = +3
Query: 143 TFTQTIFNGLNVLAGVGLLSAPDTVKQAGWASLLVIVVFAVVCFYTAELMRHCFQSREGI 202
+F T NGLN ++GVG+LS P + GW SL ++ A FY+ LM+ C + I
Sbjct: 165 SFFHTCVNGLNAISGVGILSVPYALASGGWLSLALLFCIAAAAFYSGILMKRCMEKNSNI 344
Query: 203 ISYPDIGEAAFGKYGRVFISIVLYTELYSYCVEFIIMEGDNLSGLFPGTSLHWGSLNLDG 262
+YPDIGE AFGK GR+ +SI +YTELY + F+I+EGDNLS LFP L++
Sbjct: 345 KTYPDIGELAFGKIGRLIVSISMYTELYLVSIGFLILEGDNLSNLFPIEEFQVFGLSIGA 524
Query: 263 KHLFAILAALIILPTVWLKDLR--FVSYL 289
K F IL A+IILPT+WL + F+ YL
Sbjct: 525 KKFFVILVAVIILPTIWLDNFESAFLWYL 611
>TC93131 similar to GP|19310729|gb|AAL85095.1 unknown protein {Arabidopsis
thaliana}, partial (22%)
Length = 786
Score = 77.0 bits (188), Expect = 1e-14
Identities = 40/49 (81%), Positives = 42/49 (85%)
Frame = +3
Query: 411 HML***THWQEVWRNCCLIASHALIGASFFSEQHWLYLQFVLLF*FHFS 459
+ML***THWQEV R+CCLIAS ALIGASF EQHWLYLQ VLL *F FS
Sbjct: 39 NML***THWQEV*RSCCLIASQALIGASFCLEQHWLYLQSVLLL*FLFS 185
>TC77084 similar to GP|15529155|gb|AAK97672.1 AT3g30390/T6J22_16
{Arabidopsis thaliana}, partial (88%)
Length = 2353
Score = 59.7 bits (143), Expect = 2e-09
Identities = 74/307 (24%), Positives = 125/307 (40%), Gaps = 33/307 (10%)
Frame = +1
Query: 125 KASIQKNIP-----EELPIGY----GCTFTQTIFNGLNVLAGVGLLSAPDTVKQAGWA-S 174
KA + +N P +E G+ G +F+ +FN + G G+++ P T+KQ G
Sbjct: 394 KAVVSENAPLLPKSQESDSGFDDFNGASFSGAVFNLATTIIGAGIMALPATLKQLGLIPG 573
Query: 175 LLVIVVFAVVCFYTAELMRHCFQSREGIISYPDIGEAAFGKYGRVFISIVLYTELYSYCV 234
L I++ A + + EL+ F +SY + +FGKYG+ I + +
Sbjct: 574 LCAILLMAFLTEKSIELLIR-FTRAGKAVSYAGLMGDSFGKYGKAMAQICVIVNNIGVLI 750
Query: 235 EFIIMEGDNLSGLFPGTSLHWGSL-NLDGKHLFAILAALIILPTV----------WLKDL 283
++I+ GD LSG H+G L G H + +++L TV + L
Sbjct: 751 VYMIIIGDVLSGTSSSGEHHYGILEGWFGVHWWTGRTFVVLLTTVAIFTPLASFKRIDSL 930
Query: 284 RFVSYLSAGGVVGTALVGACVYAVGTRKDVGFHHTAP-----------LVN-WSGVPFAF 331
RF S LS V ++ + V K + T P +VN ++ VP
Sbjct: 931 RFTSALSVALAVVFLVIAVGISIV---KIISGGITMPRLFPAVTDATSIVNLFTVVPVFV 1101
Query: 332 GIYGFCFAGHSVFPNIYQSMANKKDFTKAMLICFVLPVFLYGSVGAAGFLMFGERTSSQI 391
Y + HS+ + + + A+ +C +Y + GFL+FGE T +
Sbjct: 1102TAYICHYNVHSIDNELEDNSQMQGVVRTALGLC----SSVYLMISFFGFLLFGEGTLDDV 1269
Query: 392 TLDLPRD 398
+ D
Sbjct: 1270LANFDAD 1290
>BI269224 weakly similar to GP|11862972|db hypothetical protein {Oryza sativa
(japonica cultivar-group)}, partial (26%)
Length = 537
Score = 50.8 bits (120), Expect = 1e-06
Identities = 32/116 (27%), Positives = 55/116 (46%)
Frame = +2
Query: 143 TFTQTIFNGLNVLAGVGLLSAPDTVKQAGWASLLVIVVFAVVCFYTAELMRHCFQSREGI 202
+F+ ++FN + G G+++ P +K G + ++F + +T+ + F
Sbjct: 143 SFSGSVFNLSTTIIGAGIMALPAAMKVLGLTIGIASIIFLALLSHTSLDILMRFSRVAKA 322
Query: 203 ISYPDIGEAAFGKYGRVFISIVLYTELYSYCVEFIIMEGDNLSGLFPGTSLHWGSL 258
SY D+ AFG GR+ I + + V +II+ GD LSG S H+G L
Sbjct: 323 QSYGDVMGYAFGSLGRLLFQISVLFNNFGILVVYIIIIGDVLSGTTSSGSHHFGVL 490
>TC85017 weakly similar to GP|9758059|dbj|BAB08638.1 amino acid
transporter-like protein {Arabidopsis thaliana}, partial
(11%)
Length = 481
Score = 49.3 bits (116), Expect = 3e-06
Identities = 35/102 (34%), Positives = 53/102 (51%), Gaps = 2/102 (1%)
Frame = +1
Query: 128 IQKN-IPEELPIGYGCTFTQTIFNGLNVLAGVGLLSAPDTVKQAG-WASLLVIVVFAVVC 185
+Q+N IP+ PI T + +FN + G G++S P T+K G LLVIV+ AV+
Sbjct: 172 LQENPIPD--PIPQNGTVSGAVFNISTTMVGAGIMSIPATIKVLGIIPGLLVIVLVAVIT 345
Query: 186 FYTAELMRHCFQSREGIISYPDIGEAAFGKYGRVFISIVLYT 227
T E M C S + + +GE +FG G + + I + T
Sbjct: 346 DLTVEFMLRCTSSGKAVTYAGMVGE-SFGSVGSLAVKICVIT 468
>TC88142 weakly similar to GP|14587360|dbj|BAB61261. contains ESTs
AU163711(E4375) C25327(S5952) D40001(S1713)~similar to
Oryza sativa chromosome 6, partial (23%)
Length = 788
Score = 45.1 bits (105), Expect = 6e-05
Identities = 44/167 (26%), Positives = 72/167 (42%), Gaps = 11/167 (6%)
Frame = +1
Query: 148 IFNGLNVLAGVGLLSAPDTVKQAG-WASLLVIVVFAVVCFYTAELMRHCFQSREGIISYP 206
+FN + G G++S P +K G S +I++ AV+ + + + F +Y
Sbjct: 148 VFNVATSIIGSGIMSIPAILKVLGVLPSFALILIVAVLAEVSVDFLMR-FTHAGVTTTYS 324
Query: 207 DIGEAAFGKYGRVFISIVLYTELYSYCVEFIIMEGDNLSGLFPGTSLHWGSL-------- 258
+ + AFG G + + + + V F+I+ GD LSG G +H G L
Sbjct: 325 GVMKEAFGSVGALSAQVCVIITNFGGLVLFLIIIGDVLSGKESGGEVHLGLLQQWFGIHW 504
Query: 259 -NLDGKHLFAILAALIILPTVWLKDLRFVSYLSA-GGVVGTALVGAC 303
N LF I L++LP V K + + Y S ++ A V C
Sbjct: 505 WNSREVALF-ITLVLVMLPLVLYKRVESLKYSSGISTLLAVAFVTIC 642
>BF003758 similar to GP|15216030|em amino acid permease AAP4 {Vicia faba var.
minor}, partial (31%)
Length = 554
Score = 41.6 bits (96), Expect = 6e-04
Identities = 25/93 (26%), Positives = 46/93 (48%), Gaps = 6/93 (6%)
Frame = +2
Query: 152 LNVLAGVGLLSAPDTVKQAGW-ASLLVIVVFAVVCFYTAELMRHCFQSREGI-----ISY 205
+ + G G+LS + Q GW A +V+++FA V +YT+ L+ C+++ + I +Y
Sbjct: 233 ITAVIGSGVLSLAWAIAQLGWIAGPIVMILFAWVTYYTSILLCECYRTGDPISGKRNYTY 412
Query: 206 PDIGEAAFGKYGRVFISIVLYTELYSYCVEFII 238
D+ + G + IV Y L + + I
Sbjct: 413 MDVVHSNLGGFQVTLCGIVQYLNLVGVAIGYTI 511
>TC81562 weakly similar to GP|6671946|gb|AAF23206.1| putative amino acid
transporter protein {Arabidopsis thaliana}, partial
(36%)
Length = 673
Score = 40.4 bits (93), Expect = 0.001
Identities = 45/176 (25%), Positives = 70/176 (39%), Gaps = 10/176 (5%)
Frame = +1
Query: 175 LLVIVVFAVVCFYTAELMRHCFQSREGII---SYPDIGEAAFGKYGRVFISIVLYTELYS 231
L V ++F V C R S E ++ SY D+G +G GR ++ +
Sbjct: 172 LSVFILFTVWC-------RDKLASEEPLVESRSYGDLGYRCYGILGRYLTEFIIVVAQCA 330
Query: 232 YCVEFIIMEGDNLSGLFP--GTSLHWGSLNLDGKHLFAILAALIILPTVWLKDLRFVSYL 289
+ +++ G NL +F G SL ++F ++ I + W+ L ++
Sbjct: 331 GAIAYLVFIGQNLDSVFQKYGPSL--------ASYIFMLVPVEIGMS--WIGSLSALAPF 480
Query: 290 SAGGVVGTALVGACVYA--VGTRKDVGF---HHTAPLVNWSGVPFAFGIYGFCFAG 340
S V L V V + GF TA N G+PFA G+ FCF G
Sbjct: 481 SIFADVCNVLAMGIVVKEDVQVALEDGFSFRERTAITSNIGGLPFAAGMAVFCFEG 648
>TC82881 similar to GP|15216026|emb|CAC51423. amino acid permease AAP1
{Vicia faba var. minor}, partial (28%)
Length = 1132
Score = 39.3 bits (90), Expect = 0.003
Identities = 21/91 (23%), Positives = 42/91 (46%), Gaps = 6/91 (6%)
Frame = +2
Query: 152 LNVLAGVGLLSAPDTVKQAGWA-SLLVIVVFAVVCFYTAELMRHCFQSRE-----GIISY 205
+ + G G+LS + Q GW V++ F+++ +YT+ L+ C++ + ++
Sbjct: 785 ITAVVGSGVLSLAWAIAQLGWVIGPSVMIFFSLITWYTSSLLAECYRIGDPHYGKRNYTF 964
Query: 206 PDIGEAAFGKYGRVFISIVLYTELYSYCVEF 236
+ G G + IV YT LY + +
Sbjct: 965 MEAGHTILGGFNDTLCGIVQYTNLYGTAIGY 1057
>TC86488 similar to GP|9665144|gb|AAF97328.1| Unknown protein {Arabidopsis
thaliana}, partial (74%)
Length = 1753
Score = 35.4 bits (80), Expect = 0.046
Identities = 28/86 (32%), Positives = 41/86 (47%)
Frame = +1
Query: 38 NNNDDDDEDDRIATHRPESFTSQQWPQSYNEALDPLTIAAAPNIGSVLRAPSVIYASFAA 97
NN+ D + + PES SQQ + +DPL ++AAP+ S+++ AA
Sbjct: 307 NNSSISDSKEML----PESNPSQQL-STKPVPIDPLLVSAAPS--------SLVFLKAAA 447
Query: 98 GRSSKSYLELQDGFLTGTQIQESTWW 123
S S LELQ G + E TW+
Sbjct: 448 NISLASVLELQLGMFSWFSTPEVTWF 525
>TC93112 weakly similar to GP|6671946|gb|AAF23206.1| putative amino acid
transporter protein {Arabidopsis thaliana}, partial
(39%)
Length = 680
Score = 34.7 bits (78), Expect = 0.079
Identities = 18/48 (37%), Positives = 25/48 (51%)
Frame = +3
Query: 350 SMANKKDFTKAMLICFVLPVFLYGSVGAAGFLMFGERTSSQITLDLPR 397
SM +K F K + F +Y G G++ FGE T +TL+LPR
Sbjct: 39 SMKDKAKFPKLLAQTFSGITLVYILFGLCGYMAFGEETRDIVTLNLPR 182
>TC85559 similar to PIR|T07012|T07012 acetyl-CoA carboxylase (EC 6.4.1.2) -
potato chloroplast, partial (66%)
Length = 5011
Score = 34.7 bits (78), Expect = 0.079
Identities = 27/72 (37%), Positives = 40/72 (55%), Gaps = 5/72 (6%)
Frame = +3
Query: 5 NSDKNKEKENDYFLDANEDETDI-EAVNYD----SDSSNNNDDDDEDDRIATHRPESFTS 59
+SDK K+ ND D + +DI E VNY SDS N++DD+D DD T + + T
Sbjct: 2706 DSDKEKDSGNDD--DKLQKASDILEPVNYSAENYSDSDNDSDDNDPDDDYDTLQKGTDTL 2879
Query: 60 QQWPQSYNEALD 71
+ P++ N+ D
Sbjct: 2880 E--PENDNDTDD 2909
>BF632358 weakly similar to GP|19352296|gb MULTIPLE BANDED ANTIGEN
(FRAGMENT). 6/101 {Dictyostelium discoideum}, partial
(3%)
Length = 578
Score = 33.5 bits (75), Expect = 0.18
Identities = 14/43 (32%), Positives = 28/43 (64%), Gaps = 6/43 (13%)
Frame = +2
Query: 10 KEKENDYFLDANEDETDIEAVNYD------SDSSNNNDDDDED 46
+++EN+ ++ E+E D + V+YD ++ N++DDDD+D
Sbjct: 401 EDEENNNMMEDEEEEKDSDDVDYDDNDDHDAEEGNDSDDDDDD 529
>TC88143 weakly similar to GP|20453045|gb|AAM19768.1 AT3g56200/F18O21_160
{Arabidopsis thaliana}, partial (51%)
Length = 892
Score = 33.1 bits (74), Expect = 0.23
Identities = 21/84 (25%), Positives = 41/84 (48%)
Frame = +1
Query: 324 WSGVPFAFGIYGFCFAGHSVFPNIYQSMANKKDFTKAMLICFVLPVFLYGSVGAAGFLMF 383
++ VP + F F H + + K A++ C V+ Y ++G G+L+F
Sbjct: 325 FTAVPVVVTAFTFHFNVHPIGFELASPSHMKTAVRLAIMFCAVV----YFTIGLFGYLLF 492
Query: 384 GERTSSQITLDLPRDAFASKVSLW 407
G+ T S I ++ ++A ++ SL+
Sbjct: 493 GDSTQSDILINFDQNADSAVGSLF 564
>TC82248 similar to GP|15216028|emb|CAC51424. amino acid permease AAP3
{Vicia faba var. minor}, partial (29%)
Length = 779
Score = 33.1 bits (74), Expect = 0.23
Identities = 20/83 (24%), Positives = 37/83 (44%), Gaps = 1/83 (1%)
Frame = +3
Query: 143 TFTQTIFNGLNVLAGVGLLSAPDTVKQAGW-ASLLVIVVFAVVCFYTAELMRHCFQSREG 201
T T + + + G G+LS ++ Q GW A ++ F+++ YT+ + C++
Sbjct: 465 TIWTTCSHIITAVIGSGVLSLAWSIAQMGWVAGPGAMIFFSIITLYTSSFLADCYR---- 632
Query: 202 IISYPDIGEAAFGKYGRVFISIV 224
G+ FGK F+ V
Sbjct: 633 ------CGDTEFGKRNYTFMDAV 683
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.327 0.141 0.449
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,683,679
Number of Sequences: 36976
Number of extensions: 234786
Number of successful extensions: 2264
Number of sequences better than 10.0: 92
Number of HSP's better than 10.0 without gapping: 1815
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2047
length of query: 459
length of database: 9,014,727
effective HSP length: 99
effective length of query: 360
effective length of database: 5,354,103
effective search space: 1927477080
effective search space used: 1927477080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC122169.1


