

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122166.6 - phase: 0 /pseudo
(1091 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
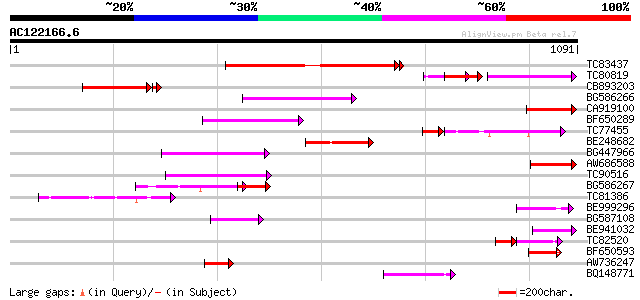
Score E
Sequences producing significant alignments: (bits) Value
TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imp... 360 7e-99
TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non... 152 1e-54
CB893203 161 8e-42
BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-li... 155 8e-38
CA919100 homologue to PIR|G90291|G902 endoglucanase precursor [i... 135 9e-32
BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgari... 134 2e-31
TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarot... 104 8e-31
BE248682 similar to GP|18568269|gb putative gag-pol polyprotein ... 129 5e-30
BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [impo... 125 7e-29
AW686588 114 2e-25
TC90516 101 2e-21
BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At... 93 6e-19
TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR re... 84 3e-16
BE999296 77 3e-14
BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - ... 75 1e-13
BE941032 75 2e-13
TC82520 56 1e-12
BF650593 71 2e-12
AW736247 weakly similar to PIR|T14619|T146 reverse transcriptase... 71 3e-12
BQ148771 70 4e-12
>TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 951
Score = 360 bits (923), Expect(2) = 7e-99
Identities = 202/335 (60%), Positives = 236/335 (70%), Gaps = 1/335 (0%)
Frame = +2
Query: 416 RFVTEFHRNRKLSKGINSRFIALIPKVASPQSLNEFRPISLVGSLYKILAKILANRLRQV 475
RFV+EFHRNRKL KGINS FIALIPKV +PQ LN+FRPISLVGSLYKIL K+LANRLR V
Sbjct: 5 RFVSEFHRNRKLFKGINSTFIALIPKVDNPQRLNDFRPISLVGSLYKILGKLLANRLRVV 184
Query: 476 IGSVVSEVQSAFVKNRQILDGILITNEVVDEARKSKKDLLLFKVDFEKAYDSVDWGYLDE 535
IGSV+S+ QSAFVKNRQIL+ + + + ++ + + K ++ G +
Sbjct: 185 IGSVISDAQSAFVKNRQILEMVFL*QMRLWMRLRN*RKIFCCLRWILKRLITLSIGLIWI 364
Query: 536 VM-GSMTFPTVW*KWIKECVGTATASVLVNGCPTDEFFLERGLRQGDPLSPFLFLLAAEG 594
+ M+F +W KWIKECV TAT SVLVNG PT
Sbjct: 365 LF*VGMSFLVLWRKWIKECVSTATTSVLVNGSPT-------------------------- 466
Query: 595 LNVLMKALVDAGLFKGYSVGRADPVVVSHLQFADDTLLIGNKSWANVRALRAGLILLEAM 654
NVLMK+LV LF YS G +PVVVSHLQFA+DTLL+ K+WAN+RALRA L++ AM
Sbjct: 467 -NVLMKSLVQTQLFTRYSFGVVNPVVVSHLQFANDTLLLETKNWANIRALRAALVIF*AM 643
Query: 655 SGLKVNFHKSSLVGVNITGSWLSEAASVLGCKVGKIPFLYLGLSIGGDPRRLLFWEPVVN 714
SGLKVNFHKS LV VNI SWLSEAASVL KVGK+PFLYLG+ I G+ RRL FWEP+VN
Sbjct: 644 SGLKVNFHKSGLVCVNIAPSWLSEAASVLSWKVGKVPFLYLGMPIEGNSRRLSFWEPIVN 823
Query: 715 RIKNRLSGWQSRFLSFGGRLVLLKFVLTALPVYAL 749
RIK RL+GW SRFLSFGGRLVLLK VLT+L VYAL
Sbjct: 824 RIKARLTGWNSRFLSFGGRLVLLKSVLTSLSVYAL 928
Score = 20.8 bits (42), Expect(2) = 7e-99
Identities = 8/8 (100%), Positives = 8/8 (100%)
Frame = +1
Query: 750 SFFKAPSG 757
SFFKAPSG
Sbjct: 928 SFFKAPSG 951
>TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non-LTR
retroelement reverse transcriptase {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 1262
Score = 152 bits (384), Expect(2) = 1e-54
Identities = 75/171 (43%), Positives = 96/171 (55%)
Frame = +2
Query: 920 LGVDVGGEALRWRRRLWAWEEELVEEFXALLLTVSLQESVTDRWIWLPTQDDGYSVRGAY 979
LG V G A W RRL+ WEEE V E LL LQ++V D+W WL +GYSV+ Y
Sbjct: 395 LGWTVDGRAWEWTRRLFVWEEECVRECCILLNNFVLQDNVNDKWRWLLDPVNGYSVKVFY 574
Query: 980 DMLTSQEQPHLHQNLELIWHTQVPLKVSILAWRLLRDRLPTKDNLANRGILPLEARMCVS 1039
+TS ++ +WH +P KVS+ WRLLR+RLPTKDNL +RG+L CV
Sbjct: 575 RYITSTGHISDRSLVDDVWHKHIPSKVSLFVWRLLRNRLPTKDNLVHRGVLLATNAACVC 754
Query: 1040 SCGNEEDVNHLFLSCATFSALWPLVQAWLGVVGVDSQSTSDHLVQFINYAG 1090
C + E HLFL C F +LW LV+ WLG+ + S H +QF G
Sbjct: 755 GCVDSESTTHLFLHCNVFCSLWSLVRNWLGIPSMSSGELRTHFIQFAKMVG 907
Score = 80.5 bits (197), Expect(2) = 1e-54
Identities = 34/73 (46%), Positives = 47/73 (63%)
Frame = +3
Query: 838 GGLEDGGRSCSSWWREIVRIRDGIGEGGEGWFGSCVRRRVGDGAETDFWWDCWCGDVSLC 897
G L +GGR S WW+ I ++R+G+GEG WF +R VGDG + FW+D W GDV L
Sbjct: 147 GWLCEGGRQSSMWWKTICKVREGVGEGVGNWFEENIRMVVGDGRDAFFWYDTWAGDVPLR 326
Query: 898 ERFSRLYDLTVNK 910
++ RL DL ++K
Sbjct: 327 LKYPRLLDLAMDK 365
Score = 65.1 bits (157), Expect = 1e-10
Identities = 41/95 (43%), Positives = 50/95 (52%), Gaps = 3/95 (3%)
Frame = +1
Query: 796 GLGVTRLREFNLALLGKWCWRLLLEKEALWRKVLVARYGVADGGLEDGGRSCSSWWREIV 855
GLGV FNL+LLGKWCWRLL++KE LW +VL ARYG GG V
Sbjct: 31 GLGVGA---FNLSLLGKWCWRLLVDKEGLWHRVLKARYGEEGGG--------------FV 159
Query: 856 RIRDGIGEGGEGWFGSCVR---RRVGDGAETDFWW 887
R+ D + GG + CV+ R G G + F W
Sbjct: 160 RVVDSLLCGGR-LYARCVKV*ERGWGTGLKKTFVW 261
>CB893203
Length = 800
Score = 161 bits (408), Expect(2) = 8e-42
Identities = 72/132 (54%), Positives = 96/132 (72%)
Frame = -3
Query: 141 LSEEWCMVWPTCLQVAQLRGLSDHCPIVLTVDEKNWGPRPVRLLKCWQDMPGYSDFVREK 200
+SE WC+ WP +Q A RGLSD CPI+LT+DE NWGPR +LKCW D+PGY FV++K
Sbjct: 798 VSESWCIKWPNMIQRALFRGLSDRCPIMLTIDEGNWGPRLHHMLKCWADLPGYHLFVKDK 619
Query: 201 WGSFQVSGWGGFVLKEKLKLMKAALKEWHVANTLNLPSKIVSLKNRRAVLDSKGEEEELS 260
W SFQVS WGG+VLKEK K+++ LK+WH +T NL ++I ++ A LDSKGE+ L
Sbjct: 618 WNSFQVSRWGGYVLKEK*KMVRNNLKDWHHNHTRNLDARINDIRESIAYLDSKGEDLTLD 439
Query: 261 EEELGELHGISS 272
EE+ +LH +S+
Sbjct: 438 TEEVDDLHTLSA 403
Score = 28.5 bits (62), Expect(2) = 8e-42
Identities = 12/16 (75%), Positives = 15/16 (93%)
Frame = -1
Query: 276 SLSRLNTSICWQQSRL 291
SLS+LNTSI WQ+SR+
Sbjct: 392 SLSKLNTSIQWQKSRI 345
>BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 789
Score = 155 bits (392), Expect = 8e-38
Identities = 87/222 (39%), Positives = 130/222 (58%), Gaps = 3/222 (1%)
Frame = -3
Query: 448 LNEFRPISLVGSLYKILAKILANRLRQVIGSVVSEVQSAFVKNRQILDGILITNEVVDEA 507
++E+R I+ + YKI+AKIL+ R++ ++ S++S QSAFV R I D +LIT++++
Sbjct: 778 VSEYRTIAPCNTQYKIIAKILSKRMQPLLRSIISPSQSAFVPGRAISDNVLITHKILHYL 599
Query: 508 RKS---KKDLLLFKVDFEKAYDSVDWGYLDEVMGSMTFPTVW*KWIKECVGTATASVLVN 564
R+S K + K D KAYD + W +L EV+ + F +W WI ECV T + S L+N
Sbjct: 598 RQSGAKKHVSMAVKTDMTKAYDRIAWNFLREVLTRLGFHGIWISWIMECVSTVSYSFLIN 419
Query: 565 GCPTDEFFLERGLRQGDPLSPFLFLLAAEGLNVLMKALVDAGLFKGYSVGRADPVVVSHL 624
G P RGLRQGDPLSP+LF+L E L+ L + + G G V R P ++HL
Sbjct: 418 GGPQGRVLPSRGLRQGDPLSPYLFILCTEVLSGLCQQALRKGTLPGVKVARNCP-PINHL 242
Query: 625 QFADDTLLIGNKSWANVRALRAGLILLEAMSGLKVNFHKSSL 666
FADDT+ G + ++ L + + A SG +N KS++
Sbjct: 241 LFADDTMFFGKSNASSCAILLSIMDKYRAASGRCIN*TKSAI 116
>CA919100 homologue to PIR|G90291|G902 endoglucanase precursor [imported] -
Sulfolobus solfataricus, partial (3%)
Length = 789
Score = 135 bits (340), Expect = 9e-32
Identities = 60/96 (62%), Positives = 73/96 (75%)
Frame = -2
Query: 995 ELIWHTQVPLKVSILAWRLLRDRLPTKDNLANRGILPLEARMCVSSCGNEEDVNHLFLSC 1054
E+IWH QVPLKVS+ AWRLLRDRLPTK NL RG++P EA +CVS CG E HLFLSC
Sbjct: 590 EMIWHRQVPLKVSVFAWRLLRDRLPTKSNLIYRGVIPTEAGLCVSGCGALESAQHLFLSC 411
Query: 1055 ATFSALWPLVQAWLGVVGVDSQSTSDHLVQFINYAG 1090
+ F++LW LV+ W+G VGVD+ SDH VQF++ G
Sbjct: 410 SYFASLWSLVRDWIGFVGVDTNVLSDHFVQFVHSTG 303
>BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgaris}, partial
(69%)
Length = 616
Score = 134 bits (337), Expect = 2e-31
Identities = 71/196 (36%), Positives = 114/196 (57%), Gaps = 1/196 (0%)
Frame = +3
Query: 371 LIKPFHEEEIKAAVWDCDSYKIPGPDGINLGFIKSFWHEMKAEIVRFVTEFHRNRKLSKG 430
L F E+K A++ DS K PG DG N+ F K W+ + ++ + +F + + K
Sbjct: 12 LCSEFTAVEVKNALFSMDSSKAPGIDGYNVHFFKCSWNIIGDSVIDAILDFFKTGFMPKI 191
Query: 431 INSRFIALIPKVASPQSLNEFRPISLVGSLYKILAKILANRLRQVIGSVVSEVQSAFVKN 490
IN ++ L+PK + S+ FRPI+ +YKI++KIL +R++ V+ SVVSE QSAFVK
Sbjct: 192 INCTYVTLLPKEVNVTSVKNFRPIACCSVIYKIISKILTSRMQGVLNSVVSENQSAFVKG 371
Query: 491 RQILDGILITNEVVDE-ARKSKKDLLLFKVDFEKAYDSVDWGYLDEVMGSMTFPTVW*KW 549
R I D I++++E+V +RK + K+D KAYDS +W ++ +M + FP + W
Sbjct: 372 RVIFDNIILSHELVKSYSRKGISPRCMVKIDLXKAYDSXEWPFIKHLMLELGFPYKFVNW 551
Query: 550 IKECVGTATASVLVNG 565
+ + TA+ + NG
Sbjct: 552 VMAXLTTASYTFNXNG 599
>TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarotenoid
dioxygenase1 {Pisum sativum}, partial (43%)
Length = 1865
Score = 104 bits (260), Expect(2) = 8e-31
Identities = 79/248 (31%), Positives = 117/248 (46%), Gaps = 16/248 (6%)
Frame = -2
Query: 837 DGGLEDGGRSCSSWWREIVRIRDGIGEGGEGWFGSCVRRRVGDGAETDFWWDCWCGDVSL 896
DGG E+ S WW++++ + + G WF + R+VGDG + FW D W V L
Sbjct: 808 DGG-ENWPAYASRWWKDLMSLEE---VGRVRWFPRELIRKVGDGRSSFFWKDAWDSSVPL 641
Query: 897 CERFSRLYDLTVNKLISVRNMILLGV----DVGGEALRWR---RRL--WAWEEELVEEFX 947
E F R + ++ +G D+ E +RWR RRL + WE+E + E
Sbjct: 640 RESFPRAF--------FPYRLLKMGCGDLWDMNAEGVRWRLYWRRLELFEWEKERLLELL 485
Query: 948 ALLLTVSLQESVTDRWIWLPTQDDGYSVRGAYDMLTSQE--QPHLHQNLELI----WHTQ 1001
L V L+ D W+W P ++ +SV Y +L + + L E+I W ++
Sbjct: 484 GRLEGVVLRYWA-DIWVWKPDKEGVFSVNSCYFLLQNLRLLEDRLSYEEEVIFRELWKSK 308
Query: 1002 VPLKVSILAWRLLRDRLPTKDNLANRGILPLE-ARMCVSSCGNEEDVNHLFLSCATFSAL 1060
P KV +W L DR+PT NL R +L +E ++ CV +E V HLFL C S +
Sbjct: 307 APAKVLAFSWTLFLDRIPTMVNLGKRRLLRVEDSKRCVFCGCQDETVVHLFLHCDVISKV 128
Query: 1061 WPLVQAWL 1068
V WL
Sbjct: 127 *REVMRWL 104
Score = 48.5 bits (114), Expect(2) = 8e-31
Identities = 21/40 (52%), Positives = 29/40 (72%)
Frame = -3
Query: 795 GGLGVTRLREFNLALLGKWCWRLLLEKEALWRKVLVARYG 834
GGLGV +R N++LL KW WRLL ++ +LW++VL YG
Sbjct: 951 GGLGVRDIRLVNVSLLAKWWWRLLQDQSSLWKEVLEDIYG 832
>BE248682 similar to GP|18568269|gb putative gag-pol polyprotein {Zea mays},
partial (1%)
Length = 441
Score = 129 bits (325), Expect = 5e-30
Identities = 69/132 (52%), Positives = 90/132 (67%)
Frame = +3
Query: 569 DEFFLERGLRQGDPLSPFLFLLAAEGLNVLMKALVDAGLFKGYSVGRADPVVVSHLQFAD 628
+E ++RGL+QGDPL+PFLFLL AEG++ LMK V+ LF+G+ V R VSHLQ+AD
Sbjct: 6 EEISVQRGLKQGDPLAPFLFLLVAEGISGLMKNAVNRNLFQGFDVKRGG-TRVSHLQYAD 182
Query: 629 DTLLIGNKSWANVRALRAGLILLEAMSGLKVNFHKSSLVGVNITGSWLSEAASVLGCKVG 688
DTL IG + N+ L+A L E SGLKVNFHKSSL+G+N+ ++ A L C+
Sbjct: 183 DTLCIGMPTVDNLWTLKALLQGFEMASGLKVNFHKSSLIGINVPRDFMEAACRFLNCREE 362
Query: 689 KIPFLYLGLSIG 700
IPF+YLGL G
Sbjct: 363 SIPFIYLGLPGG 398
>BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [imported] -
Arabidopsis thaliana, partial (7%)
Length = 687
Score = 125 bits (315), Expect = 7e-29
Identities = 73/209 (34%), Positives = 113/209 (53%), Gaps = 2/209 (0%)
Frame = +2
Query: 293 WLRDGDANSKFFHSVLAARRRINSLSSILVD-GVVVEGVQPVRQAVFTHSKNHFLAPIMT 351
WL+DGD N+KFFHS + RR++N + + + G +G + V + + T+ N F + T
Sbjct: 5 WLKDGDKNTKFFHSKASQRRKVNEIKKLKDETGNWCKGEENVERLLITYFNNLFTSSNPT 184
Query: 352 R-PSVGHLQFRKLSSVERGGLIKPFHEEEIKAAVWDCDSYKIPGPDGINLGFIKSFWHEM 410
+ KLS K F EEE+ A+ K PGPDG+ F + +WH +
Sbjct: 185 AIEETCEVVKGKLSHEHIVWCEKEFTEEEVLEAINQMHPVKAPGPDGLPALFFQKYWHIV 364
Query: 411 KAEIVRFVTEFHRNRKLSKGINSRFIALIPKVASPQSLNEFRPISLVGSLYKILAKILAN 470
E+ + V + N ++ +N FI LIPK +P + ++RPISL + KI+ K++AN
Sbjct: 365 GKEVQQMVLQVLNNSMETEELNKTFIVLIPKGKNPNTPKDYRPISLCNVVMKIITKVIAN 544
Query: 471 RLRQVIGSVVSEVQSAFVKNRQILDGILI 499
R++Q + V+ QSAFV+ R I D LI
Sbjct: 545 RVKQTLPDVIDVEQSAFVQGRLITDNALI 631
>AW686588
Length = 567
Score = 114 bits (286), Expect = 2e-25
Identities = 54/89 (60%), Positives = 61/89 (67%)
Frame = +1
Query: 1002 VPLKVSILAWRLLRDRLPTKDNLANRGILPLEARMCVSSCGNEEDVNHLFLSCATFSALW 1061
VPLKVSILAWRL+RDRLPTK NL R L +EA CV CG E NHLFL CATF A+W
Sbjct: 142 VPLKVSILAWRLIRDRLPTKANLVRRRCLAVEAAGCVVGCGIAETANHLFLHCATFGAVW 321
Query: 1062 PLVQAWLGVVGVDSQSTSDHLVQFINYAG 1090
++AW+GV G D SDH +QFI G
Sbjct: 322 QHIRAWIGVSGADPHDLSDHFIQFITCTG 408
>TC90516
Length = 983
Score = 101 bits (251), Expect = 2e-21
Identities = 64/205 (31%), Positives = 110/205 (53%)
Frame = -2
Query: 300 NSKFFHSVLAARRRINSLSSILVDGVVVEGVQPVRQAVFTHSKNHFLAPIMTRPSVGHLQ 359
N+ +F + + R NS+S+ V+ + + V +QA+ +HF P RP++G +
Sbjct: 742 NTTYFLACVKNRGMRNSISAPRVERWM-DDVAKNKQAIVNFFTHHFSDP*TYRPTMGDID 566
Query: 360 FRKLSSVERGGLIKPFHEEEIKAAVWDCDSYKIPGPDGINLGFIKSFWHEMKAEIVRFVT 419
F +S+++ L F +++ V D + P PDG NL F + +KA+I
Sbjct: 565 FFHISNLDNVLLSAQFLVSKMELVVSSLDGNESPRPDGFNLNFFIRLRNMLKADIEIMFE 386
Query: 420 EFHRNRKLSKGINSRFIALIPKVASPQSLNEFRPISLVGSLYKILAKILANRLRQVIGSV 479
+F+ L K + F+ LI KV P L +F +S +GSL K++AK+LA RL ++ +
Sbjct: 385 QFYTPANLLKIFS*YFLTLISKVEYPILLGDFSLMSFLGSL*KLMAKVLALRLAHIMEKI 206
Query: 480 VSEVQSAFVKNRQILDGILITNEVV 504
+ QS FV+ RQ +DG++ NE++
Sbjct: 205 IFVNQSTFVRGRQHVDGVVAINEII 131
>BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (16%)
Length = 794
Score = 92.8 bits (229), Expect = 6e-19
Identities = 73/225 (32%), Positives = 108/225 (47%), Gaps = 10/225 (4%)
Frame = +3
Query: 243 LKNRRA--VLDSKGEEEELSEEELGELHGISSDIHSLSRLNTSICW-QQSRLNWLRDGDA 299
L++RR L +KGEE L EL E + H N I W Q+SRLNWLR GD
Sbjct: 33 LQDRRGNTKLFNKGEELSLLRSELNE------EYH-----NEEIFWMQKSRLNWLRSGDR 179
Query: 300 NSKFFHSVLAARRRINSLSSILVDGVVVEGVQPVRQAVFTHSKNHFLAPIMTRPSVGHLQ 359
N+KFFH+V RR N + S++ D + V + + + +HF + + VG +
Sbjct: 180 NTKFFHAVTKNRRAQNRILSLIDDD---DKEWFVEEDLGRLADSHFKL-LYSSEDVG-IT 344
Query: 360 FRKLSSV-------ERGGLIKPFHEEEIKAAVWDCDSYKIPGPDGINLGFIKSFWHEMKA 412
+S+ + L+ EE++ AV+D + +K PGPDG+N+ F + FW M
Sbjct: 345 LEDWNSIPAIVTEEQNAQLMAQISREEVREAVFDINPHKCPGPDGMNVFFFQQFWDTMGD 524
Query: 413 EIVRFVTEFHRNRKLSKGINSRFIALIPKVASPQSLNEFRPISLV 457
++ EF R KL +GIN I K A + + P V
Sbjct: 525 DLTSMAQEFLRTGKLEEGINKTNIWFGSKKAGGKEASGISPHKFV 659
Score = 57.4 bits (137), Expect = 3e-08
Identities = 28/64 (43%), Positives = 43/64 (66%)
Frame = +1
Query: 438 LIPKVASPQSLNEFRPISLVGSLYKILAKILANRLRQVIGSVVSEVQSAFVKNRQILDGI 497
L+PK + L EFRPISL YKI++K+L+ RL+ V+ +++E Q+AF + + I D I
Sbjct: 601 LVPKKLEAKRLVEFRPISLCNVAYKIVSKVLSKRLKSVLPWIITETQAAFGRRQLISDNI 780
Query: 498 LITN 501
LI +
Sbjct: 781 LIAH 792
>TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR retroelement
reverse transcriptase {Oryza sativa (japonica
cultivar-group)}, partial (1%)
Length = 798
Score = 84.0 bits (206), Expect = 3e-16
Identities = 76/277 (27%), Positives = 134/277 (47%), Gaps = 13/277 (4%)
Frame = -1
Query: 55 KQVLWNSLSSRLLSLGSSKVCVCGDFNTVRSVDERRSVRGSQVVDDCAPFNDFIEDRVLI 114
++ LW+++S+ S C+ GDFNT+ E + + F + + L
Sbjct: 795 RRQLWSAISNIQTQHKLSWCCI-GDFNTILGSHEHQGSHTPARLP-MLDFQQWSDVNNLF 622
Query: 115 NLPLCGRKFTWYNGDGRSMST---LDRFLLSE---EWCMVWPTCLQVAQLRGLSDHCPIV 168
+LP G FTW NG +T LDR ++++ + C C + +LR SDH PI+
Sbjct: 621 HLPTRGSAFTWTNGRRGRNNTRKRLDRSIVNQLMIDNCESLSACT-LTKLR--SDHFPIL 451
Query: 169 LTVDEKNWG-PRPVRLLKCWQDMPGYSDFVREKWGSFQVSGWGGFVLKEKLKLMKAALKE 227
+ +N + +K W P + +++ W + QV G FVL +KLK +K LK
Sbjct: 450 FELQTQNIQFSSSFKFMKMWSAHPDCINIIKQCWAN-QVVGCPMFVLNQKLKNLKEVLKV 274
Query: 228 WHVANTLNLPSKIVS----LKNRRAVLDSKGEEEELSEEELGELHGISSDIHSLSRLNTS 283
W+ N+ S++ + L + + +DS G + L ++E ++ ++ S LN
Sbjct: 273 WNKNTFGNVHSQVDNAYKELDDIQVKIDSIGYSDVLMDQEK------AAQLNLESALNIE 112
Query: 284 -ICW-QQSRLNWLRDGDANSKFFHSVLAARRRINSLS 318
+ W ++S++NW +GD N+ FFH V +R + +S
Sbjct: 111 EVFWHEKSKVNWHCEGDRNTAFFHRVAKIKRTSSLIS 1
>BE999296
Length = 384
Score = 77.4 bits (189), Expect = 3e-14
Identities = 40/110 (36%), Positives = 57/110 (51%)
Frame = -1
Query: 975 VRGAYDMLTSQEQPHLHQNLELIWHTQVPLKVSILAWRLLRDRLPTKDNLANRGILPLEA 1034
+ GAY L S + P + ++ +WH +PLKV R+LR+ LPTKDN R ++ E
Sbjct: 306 IHGAYQFLMSADAPLDREYIDSVWHNHIPLKVCFFVLRVLRNCLPTKDNFVRRRVIHEEH 127
Query: 1035 RMCVSSCGNEEDVNHLFLSCATFSALWPLVQAWLGVVGVDSQSTSDHLVQ 1084
+C + C +E + LF LWPLV WL + V+S DH Q
Sbjct: 126 MLCPTGCSFKETTDDLF--------LWPLV*QWLHIS*VNSCVLRDHFYQ 1
>BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - Arabidopsis
thaliana, partial (17%)
Length = 677
Score = 75.5 bits (184), Expect = 1e-13
Identities = 40/102 (39%), Positives = 58/102 (56%)
Frame = +3
Query: 387 CDSYKIPGPDGINLGFIKSFWHEMKAEIVRFVTEFHRNRKLSKGINSRFIALIPKVASPQ 446
C K+ G L F + WH +K ++++ V F + KL +N+ I LIPK P
Sbjct: 366 CTRKKLLGRME*QLSFFQHSWHIIKMDLLKMVNSFLASGKLDTRLNTTNICLIPKKKRPT 545
Query: 447 SLNEFRPISLVGSLYKILAKILANRLRQVIGSVVSEVQSAFV 488
+ E RPISL YKI++K+L RL+ + S++SE QSAFV
Sbjct: 546 RMTELRPISLCNVGYKIISKVLCQRLKVCLPSLISETQSAFV 671
Score = 38.1 bits (87), Expect = 0.019
Identities = 33/117 (28%), Positives = 52/117 (44%), Gaps = 4/117 (3%)
Frame = +2
Query: 286 WQQ-SRLNWLRDGDANSKFFHSVLAARRRINSLSSIL-VDGVVVEGVQPVRQAVFTHSKN 343
WQQ SR W GD N KF+H++ R N + + DG + Q V + + ++
Sbjct: 53 WQQKSRNMWHISGDLNKKFYHALTKQRHARNRIVGLYDYDGNWITEEQGVEKVAVDYFED 232
Query: 344 HFLAPIMTRPSVGHLQ--FRKLSSVERGGLIKPFHEEEIKAAVWDCDSYKIPGPDGI 398
F T G L ++ L++ EEE++ A++ K PGPDG+
Sbjct: 233 LFQRTTPTGFD-GFLDEITSSITPQMNQRLLRLATEEEVRLALFIMHPEKAPGPDGM 400
>BE941032
Length = 435
Score = 74.7 bits (182), Expect = 2e-13
Identities = 37/84 (44%), Positives = 45/84 (53%)
Frame = +2
Query: 1007 SILAWRLLRDRLPTKDNLANRGILPLEARMCVSSCGNEEDVNHLFLSCATFSALWPLVQA 1066
SI W +L +R PTKDNL RG++ + CV CGN D HLFL C F +W V
Sbjct: 146 SIFLWCVLLNRFPTKDNLLKRGVISAIYQSCVGECGNLYDATHLFLHCNFFRQIWINVSD 325
Query: 1067 WLGVVGVDSQSTSDHLVQFINYAG 1090
WL V V SD L QF ++ G
Sbjct: 326 WLSFVMVTLLRISDRLAQFGSFGG 397
>TC82520
Length = 833
Score = 56.2 bits (134), Expect(2) = 1e-12
Identities = 31/89 (34%), Positives = 43/89 (47%)
Frame = +3
Query: 976 RGAYDMLTSQEQPHLHQNLELIWHTQVPLKVSILAWRLLRDRLPTKDNLANRGILPLEAR 1035
RG Y LT QP ++ +W +P KVS+ WRL +RLPTK NL R +L
Sbjct: 123 RGTYRFLTISGQPLDRNQVDDVWQKNIPSKVSMFVWRLFHNRLPTKVNLMQRHVLQQTHT 302
Query: 1036 MCVSSCGNEEDVNHLFLSCATFSALWPLV 1064
C+S + H+ F AL+ L+
Sbjct: 303 ACISGVAIRKR-QHICFYIVIFLALFGLM 386
Score = 41.2 bits (95), Expect = 0.002
Identities = 19/50 (38%), Positives = 25/50 (50%)
Frame = +2
Query: 1041 CGNEEDVNHLFLSCATFSALWPLVQAWLGVVGVDSQSTSDHLVQFINYAG 1090
CG+ E HLFL C F +LW V WL ++ V +QF + AG
Sbjct: 317 CGDSETATHLFLHCDIFGSLWSHVLRWLHLLLVLPADIRQFFIQFTSMAG 466
Score = 35.8 bits (81), Expect(2) = 1e-12
Identities = 20/40 (50%), Positives = 24/40 (60%)
Frame = +2
Query: 935 LWAWEEELVEEFXALLLTVSLQESVTDRWIWLPTQDDGYS 974
L+AWEEE V E+ ALL LQE+V D WL GY+
Sbjct: 2 LFAWEEESVREWYALLHNTVLQENVHDVCRWLLDPI*GYT 121
>BF650593
Length = 486
Score = 71.2 bits (173), Expect = 2e-12
Identities = 32/63 (50%), Positives = 40/63 (62%)
Frame = +3
Query: 999 HTQVPLKVSILAWRLLRDRLPTKDNLANRGILPLEARMCVSSCGNEEDVNHLFLSCATFS 1058
H VPLKVS L WRL ++ L T+DNL+ RG+L + CV CG EE V+H F C FS
Sbjct: 297 HKSVPLKVSCLVWRLFQNXLATRDNLSKRGVLDQNSIXCVXDCGREESVSHFFFEC-PFS 473
Query: 1059 ALW 1061
+W
Sbjct: 474 XVW 482
Score = 32.3 bits (72), Expect = 1.0
Identities = 14/37 (37%), Positives = 20/37 (53%)
Frame = +1
Query: 824 LWRKVLVARYGVADGGLEDGGRSCSSWWREIVRIRDG 860
LW + LV +YG+ G + R S WW++I I G
Sbjct: 88 LWFRALVNKYGLNRGSITIENRGVSLWWKDICYIDFG 198
>AW736247 weakly similar to PIR|T14619|T146 reverse transcriptase - beet
retrotransposon (fragment), partial (4%)
Length = 305
Score = 70.9 bits (172), Expect = 3e-12
Identities = 31/58 (53%), Positives = 39/58 (66%), Gaps = 2/58 (3%)
Frame = +1
Query: 375 FHEEEIKAAVWDCDSYKI--PGPDGINLGFIKSFWHEMKAEIVRFVTEFHRNRKLSKG 430
F EEE++ AVWDCDS PGPDG+N F+K +W +K + +R V EFH N KL KG
Sbjct: 121 FSEEEVRKAVWDCDSSHS*NPGPDGVNFTFVKEYWELIKVDFLRVVMEFHTNGKLGKG 294
>BQ148771
Length = 680
Score = 70.1 bits (170), Expect = 4e-12
Identities = 43/141 (30%), Positives = 71/141 (49%), Gaps = 3/141 (2%)
Frame = -3
Query: 720 LSGWQSRFLSFGGRLVLLKFVLTALPVYALSFFKAPSGIISSIESLFNKFFWGGGEDKRK 779
L+ W++ LS R+ L K V+ A+P+Y + P I I+ L KF WG E R+
Sbjct: 573 LANWKANHLSLARRVTLAKSVIEAVPLYPMMTTIIPKACIEEIQKLQRKFVWGDTEVSRR 394
Query: 780 ISWIRWDTLSLRKEYGGLGVTRLREFNLALLGKWCWRLLLEKEALWRKVLVARYGVADGG 839
+ W+T+S K GLG+ RL N A + K W + +L +V+ +Y ++
Sbjct: 393 YHAVGWETMSKPKTIYGLGLRRLDVMNKACIMKLGWSIYSGSNSLCTEVMRGKYQRSE-S 217
Query: 840 LED---GGRSCSSWWREIVRI 857
LE+ + SS W+ +V++
Sbjct: 216 LEEIFLEKPTDSSLWKALVKL 154
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.140 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 38,645,672
Number of Sequences: 36976
Number of extensions: 625860
Number of successful extensions: 3362
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 3274
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3342
length of query: 1091
length of database: 9,014,727
effective HSP length: 106
effective length of query: 985
effective length of database: 5,095,271
effective search space: 5018841935
effective search space used: 5018841935
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 64 (29.3 bits)
Medicago: description of AC122166.6


