

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122166.2 + phase: 0
(395 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
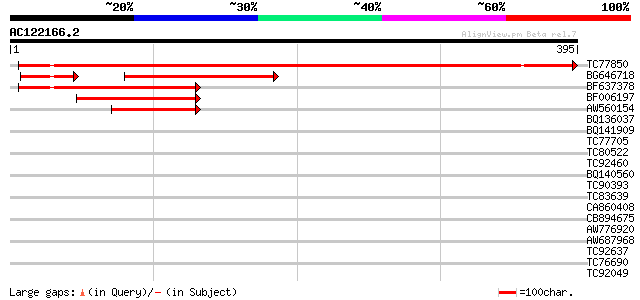
Score E
Sequences producing significant alignments: (bits) Value
TC77850 weakly similar to PIR|H96517|H96517 protein T2E6.22 [imp... 504 e-143
BG646718 weakly similar to PIR|H96517|H96 protein T2E6.22 [impor... 152 1e-44
BF637378 weakly similar to PIR|H96517|H965 protein T2E6.22 [impo... 129 2e-30
BF006197 weakly similar to GP|1310677|emb| protein z-type serpin... 93 2e-19
AW560154 76 2e-14
BQ136037 similar to GP|21322711|em pherophorin-dz1 protein {Volv... 30 1.2
BQ141909 weakly similar to GP|14253159|emb VMP3 protein {Volvox ... 24 1.5
TC77705 similar to GP|21553857|gb|AAM62950.1 unknown {Arabidopsi... 29 2.8
TC80522 weakly similar to PIR|H96548|H96548 unknown protein [imp... 29 3.6
TC92460 similar to GP|22202705|dbj|BAC07363. hypothetical protei... 29 3.6
BQ140560 weakly similar to GP|15128223|db hypothetical protein~s... 24 3.7
TC90393 similar to GP|15293299|gb|AAK93760.1 unknown protein {Ar... 28 4.7
TC83639 GP|13277222|emb|CAC34410. ZF-HD homeobox protein {Flaver... 25 5.4
CA860408 GP|10728858|g CG10600 gene product {Drosophila melanoga... 28 6.2
CB894675 28 6.2
AW776920 similar to PIR|S47243|S4 starch phosphorylase (EC 2.4.1... 28 8.0
AW687968 similar to GP|22136982|gb| unknown protein {Arabidopsis... 28 8.0
TC92637 similar to PIR|T39903|T39903 serine-rich protein - fissi... 28 8.0
TC76690 similar to GP|21593806|gb|AAM65773.1 putative RING zinc ... 28 8.0
TC92049 similar to PIR|T01061|T01061 hypothetical protein YUP8H1... 23 8.5
>TC77850 weakly similar to PIR|H96517|H96517 protein T2E6.22 [imported] -
Arabidopsis thaliana, partial (65%)
Length = 1485
Score = 504 bits (1299), Expect = e-143
Identities = 254/389 (65%), Positives = 313/389 (80%)
Frame = +2
Query: 7 KSFTNLTNISMNITKHLLSNQKLKEENVVFSPLSLNTVLSMIATGSEGPTQKQLLSFLQS 66
+S N TN+S+++ KHL S + + N+VFSPLSL VLS+IA+GSEGPTQ+QL +FLQS
Sbjct: 53 ESIANQTNVSLSVAKHLFSKES--DNNIVFSPLSLQVVLSIIASGSEGPTQQQLFNFLQS 226
Query: 67 ESTGDLKSLCSQLVSSVLSDGAPAGGPCLSYVNGVWVEQTIPLQPSFKQLMNTDFKAAFA 126
+ST L SQLVS +LSD +PAGGP LS+V+GVWV+QT+ LQPSF+Q+++T FKAA +
Sbjct: 227 KSTDHLNYFASQLVSVILSDASPAGGPLLSFVDGVWVDQTLSLQPSFQQIVSTHFKAALS 406
Query: 127 AVDFVNKANEVGEEVNFWAEKETKGLIKNLLPPGSVNSLTRLIFANALYFKGVWKQQFDT 186
+VDF NKA EV EVN WAEKET GLIK LLP GSVN+ TRLIFANALYFKG W +FD
Sbjct: 407 SVDFQNKAVEVTNEVNSWAEKETNGLIKELLPLGSVNNATRLIFANALYFKGAWNDKFDA 586
Query: 187 TKTKDYDFDLLNGKSVKVPFMTSKNDQFISSLDGFKVLGLPYKQGKDERAFSIYFFLPDK 246
+KT+DY+F LLNG VKVPFMTSK QFI + DGFKVLGLPYKQG+D+R F++YFFLP+
Sbjct: 587 SKTEDYEFHLLNGSPVKVPFMTSKKKQFIRAFDGFKVLGLPYKQGEDKRQFTMYFFLPNA 766
Query: 247 KDGLSNLIDKVASDSEFLERNLPRRKVEVGKFRIPRFNISFEIEASELLKKLGLALPFTL 306
KDGL+ L++KVAS+SE L+ LP KVEVG FRIP+FNISF +E S++LK+LG+ LPF+
Sbjct: 767 KDGLAALVEKVASESELLQHKLPFGKVEVGDFRIPKFNISFGLETSDMLKELGVVLPFSG 946
Query: 307 GGLTKMVDSPISQELYVSGIFQKSFIEVNEEGTKAAAVTVSFISSRSRYSPPPPPPIDFV 366
GGLTKMV+S +SQ L VS IF KSFIEVNEEGT+AAA T + I RS S PP +DFV
Sbjct: 947 GGLTKMVNSSVSQNLCVSNIFHKSFIEVNEEGTEAAAATAATILLRSAMS--IPPRLDFV 1120
Query: 367 ADHPFLFLIREEFSGTILFVGKVVNPLDG 395
ADHPFLF+IRE+ +GTI+FVG+V+NPL G
Sbjct: 1121ADHPFLFMIREDLTGTIIFVGQVLNPLAG 1207
>BG646718 weakly similar to PIR|H96517|H96 protein T2E6.22 [imported] -
Arabidopsis thaliana, partial (30%)
Length = 702
Score = 152 bits (384), Expect(2) = 1e-44
Identities = 71/107 (66%), Positives = 89/107 (82%)
Frame = +2
Query: 81 SSVLSDGAPAGGPCLSYVNGVWVEQTIPLQPSFKQLMNTDFKAAFAAVDFVNKANEVGEE 140
S VLSD A AGGP LS+V+GVW+E+T+ LQPSFKQ+M++D+KA A+VDF+ KA EV +E
Sbjct: 224 SIVLSDAASAGGPHLSFVDGVWIEKTLSLQPSFKQIMSSDYKATLASVDFMFKAPEVRKE 403
Query: 141 VNFWAEKETKGLIKNLLPPGSVNSLTRLIFANALYFKGVWKQQFDTT 187
VN W EKET GLIK L+PPGSV+SLT LIFANALYF+G W ++FD +
Sbjct: 404 VNIWVEKETNGLIKELIPPGSVDSLTSLIFANALYFRGAWNEKFDVS 544
Score = 45.4 bits (106), Expect(2) = 1e-44
Identities = 23/41 (56%), Positives = 28/41 (68%)
Frame = +1
Query: 8 SFTNLTNISMNITKHLLSNQKLKEENVVFSPLSLNTVLSMI 48
S TN T +S+ I KHL S Q E N+VFSPLSL VLS++
Sbjct: 22 SMTNQTRVSLRIAKHLFSKQS--ENNIVFSPLSLQVVLSIL 138
Score = 36.2 bits (82), Expect = 0.023
Identities = 21/42 (50%), Positives = 27/42 (64%)
Frame = +3
Query: 44 VLSMIATGSEGPTQKQLLSFLQSESTGDLKSLCSQLVSSVLS 85
V + A S+ PT +QLL FL+S+ST L L SQLVS + S
Sbjct: 111 VSASCAQHSDCPTPQQLLDFLRSKSTDHLNYLASQLVSLLCS 236
>BF637378 weakly similar to PIR|H96517|H965 protein T2E6.22 [imported] -
Arabidopsis thaliana, partial (9%)
Length = 627
Score = 129 bits (324), Expect = 2e-30
Identities = 66/127 (51%), Positives = 89/127 (69%)
Frame = +1
Query: 7 KSFTNLTNISMNITKHLLSNQKLKEENVVFSPLSLNTVLSMIATGSEGPTQKQLLSFLQS 66
+S N T +S+ I KHL S + E+N+VFSPLSL VLS+I GSEG TQ+QLL FL+
Sbjct: 19 ESSNNQTKVSLTIVKHLFSKES--EKNIVFSPLSLQVVLSIITAGSEGSTQQQLLDFLRF 192
Query: 67 ESTGDLKSLCSQLVSSVLSDGAPAGGPCLSYVNGVWVEQTIPLQPSFKQLMNTDFKAAFA 126
+ST L S S +S +L D +GGP LS+V+GVW+E+T+ LQ SFKQ+++ D+KA +
Sbjct: 193 KSTDHLNSSVSHQLSIILKDATSSGGPHLSFVDGVWLEKTLSLQSSFKQIVSEDYKATLS 372
Query: 127 AVDFVNK 133
VDF NK
Sbjct: 373 EVDFKNK 393
>BF006197 weakly similar to GP|1310677|emb| protein z-type serpin {Hordeum
vulgare subsp. vulgare}, partial (7%)
Length = 468
Score = 92.8 bits (229), Expect = 2e-19
Identities = 47/87 (54%), Positives = 59/87 (67%)
Frame = +3
Query: 47 MIATGSEGPTQKQLLSFLQSESTGDLKSLCSQLVSSVLSDGAPAGGPCLSYVNGVWVEQT 106
++ EG TQ+QLL FL+S+ L S S LVS +LSD AP+GG LS VWV+QT
Sbjct: 3 IVTAACEGRTQQQLLEFLRSKFIDHLNSFTSHLVSIILSDAAPSGGSRLSLTQRVWVDQT 182
Query: 107 IPLQPSFKQLMNTDFKAAFAAVDFVNK 133
+ LQPSFK+ M TD+KA A+VDF NK
Sbjct: 183 LSLQPSFKETMVTDYKATLASVDFQNK 263
>AW560154
Length = 470
Score = 76.3 bits (186), Expect = 2e-14
Identities = 37/62 (59%), Positives = 45/62 (71%)
Frame = +1
Query: 72 LKSLCSQLVSSVLSDGAPAGGPCLSYVNGVWVEQTIPLQPSFKQLMNTDFKAAFAAVDFV 131
L S S LVS +LSD AP+GG LS+ VWV+QT+ LQPSFK+ M TD+KA A+VDF
Sbjct: 7 LNSFTSDLVSIILSDAAPSGGSRLSFTQRVWVDQTLSLQPSFKETMVTDYKATLASVDFQ 186
Query: 132 NK 133
NK
Sbjct: 187 NK 192
>BQ136037 similar to GP|21322711|em pherophorin-dz1 protein {Volvox carteri
f. nagariensis}, partial (5%)
Length = 712
Score = 30.4 bits (67), Expect = 1.2
Identities = 13/21 (61%), Positives = 14/21 (65%)
Frame = +2
Query: 345 TVSFISSRSRYSPPPPPPIDF 365
+V F SR PPPPPPIDF
Sbjct: 305 SVFFFFSRHLPPPPPPPPIDF 367
>BQ141909 weakly similar to GP|14253159|emb VMP3 protein {Volvox carteri f.
nagariensis}, partial (13%)
Length = 1223
Score = 24.3 bits (51), Expect(2) = 1.5
Identities = 9/15 (60%), Positives = 9/15 (60%)
Frame = -1
Query: 357 PPPPPPIDFVADHPF 371
PPPPPP HPF
Sbjct: 209 PPPPPPPPASPFHPF 165
Score = 24.3 bits (51), Expect(2) = 1.5
Identities = 10/16 (62%), Positives = 10/16 (62%)
Frame = -2
Query: 347 SFISSRSRYSPPPPPP 362
SF SS PPPPPP
Sbjct: 316 SFCSSPPFLPPPPPPP 269
>TC77705 similar to GP|21553857|gb|AAM62950.1 unknown {Arabidopsis
thaliana}, partial (58%)
Length = 1139
Score = 29.3 bits (64), Expect = 2.8
Identities = 30/93 (32%), Positives = 46/93 (49%), Gaps = 8/93 (8%)
Frame = +1
Query: 28 KLKEENVVFSP-LSLNTVLSM-IATGSEGPTQKQLLSFLQSESTGDLKSLCSQLV--SSV 83
+ K + + +P +S T LS + T S+ PTQ +L S +QSES K+ S + SV
Sbjct: 682 EFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSSSIQTESV 861
Query: 84 LSDGA----PAGGPCLSYVNGVWVEQTIPLQPS 112
+D + P P + V+ V VE I + S
Sbjct: 862 ATDASISVPPESSP--AEVSQVQVESQIQTETS 954
>TC80522 weakly similar to PIR|H96548|H96548 unknown protein [imported] -
Arabidopsis thaliana, partial (12%)
Length = 807
Score = 28.9 bits (63), Expect = 3.6
Identities = 11/30 (36%), Positives = 17/30 (56%)
Frame = -1
Query: 334 VNEEGTKAAAVTVSFISSRSRYSPPPPPPI 363
VNE T++ + ++ SPPPPPP+
Sbjct: 456 VNESTTESVIGAIEAVTETLSASPPPPPPL 367
>TC92460 similar to GP|22202705|dbj|BAC07363. hypothetical protein {Oryza
sativa (japonica cultivar-group)}, partial (9%)
Length = 795
Score = 28.9 bits (63), Expect = 3.6
Identities = 25/88 (28%), Positives = 33/88 (37%), Gaps = 5/88 (5%)
Frame = +3
Query: 282 RFNISFEIEASELLKKLGLALPFTLGGLTKMVDS-----PISQELYVSGIFQKSFIEVNE 336
R + E EA+ KK L+ T T S P + L G K+ ++ E
Sbjct: 501 RISFPSEEEAAAAAKKRKLSASSTTEASTAATASVSAKAPSTTHLSNGGRKSKAVMDDYE 680
Query: 337 EGTKAAAVTVSFISSRSRYSPPPPPPID 364
F SRY P PPPP+D
Sbjct: 681 SSDDDEDDERHFKRRPSRYEPSPPPPVD 764
>BQ140560 weakly similar to GP|15128223|db hypothetical protein~similar to
Oryza sativa chromosome 1 P0489A01.2, partial (5%)
Length = 215
Score = 24.3 bits (51), Expect(2) = 3.7
Identities = 9/18 (50%), Positives = 10/18 (55%)
Frame = +1
Query: 357 PPPPPPIDFVADHPFLFL 374
PPPPPP P L+L
Sbjct: 133 PPPPPPFHGAPPPPXLYL 186
Score = 23.1 bits (48), Expect(2) = 3.7
Identities = 8/12 (66%), Positives = 9/12 (74%)
Frame = +1
Query: 351 SRSRYSPPPPPP 362
S S+ PPPPPP
Sbjct: 1 SMSKAPPPPPPP 36
>TC90393 similar to GP|15293299|gb|AAK93760.1 unknown protein {Arabidopsis
thaliana}, partial (14%)
Length = 754
Score = 28.5 bits (62), Expect = 4.7
Identities = 23/68 (33%), Positives = 36/68 (52%), Gaps = 4/68 (5%)
Frame = +3
Query: 23 LLSNQKLKEENVVFSPLSLNTVLSMIATGSEGPTQKQLLSFLQSESTGDL-KSLC---SQ 78
LL Q ++++ ++ LS NT S+ + LLS +QSE+ + K LC S+
Sbjct: 435 LLRKQLTRDDSFLWPRLSSNTQASL---------KSLLLSSIQSENAKSISKKLCDTISE 587
Query: 79 LVSSVLSD 86
L SS+L D
Sbjct: 588 LASSILPD 611
>TC83639 GP|13277222|emb|CAC34410. ZF-HD homeobox protein {Flaveria
bidentis}, partial (4%)
Length = 670
Score = 25.4 bits (54), Expect(2) = 5.4
Identities = 11/22 (50%), Positives = 12/22 (54%)
Frame = -1
Query: 357 PPPPPPIDFVADHPFLFLIREE 378
PPPPPP V P L L + E
Sbjct: 292 PPPPPPASCVMKFPPLMLPKAE 227
Score = 21.2 bits (43), Expect(2) = 5.4
Identities = 8/19 (42%), Positives = 11/19 (57%)
Frame = -1
Query: 344 VTVSFISSRSRYSPPPPPP 362
+ +SFI + PPPPP
Sbjct: 337 IPLSFIVNLHEPPSPPPPP 281
>CA860408 GP|10728858|g CG10600 gene product {Drosophila melanogaster},
partial (0%)
Length = 451
Score = 28.1 bits (61), Expect = 6.2
Identities = 20/62 (32%), Positives = 30/62 (48%), Gaps = 3/62 (4%)
Frame = +2
Query: 322 YVSGIFQKSFIEVNEEGTKAAAVTVSFISSRSRY---SPPPPPPIDFVADHPFLFLIREE 378
+VS IF++ V+ + ++ + S IS S +PPPPPP F D L +
Sbjct: 77 HVSEIFRRPHGGVSPFSSLSSLWSSSSISRHSSTRTSAPPPPPPPPFKPDQSVLLVGEGN 256
Query: 379 FS 380
FS
Sbjct: 257 FS 262
>CB894675
Length = 167
Score = 28.1 bits (61), Expect = 6.2
Identities = 10/20 (50%), Positives = 14/20 (70%)
Frame = +1
Query: 352 RSRYSPPPPPPIDFVADHPF 371
R +SPPPPPP D + +P+
Sbjct: 76 RPPWSPPPPPPPDHKSTNPY 135
>AW776920 similar to PIR|S47243|S4 starch phosphorylase (EC 2.4.1.1) isoform
L precursor chloroplast - fava bean, partial (17%)
Length = 546
Score = 27.7 bits (60), Expect = 8.0
Identities = 12/32 (37%), Positives = 18/32 (55%)
Frame = -1
Query: 36 FSPLSLNTVLSMIATGSEGPTQKQLLSFLQSE 67
++PL LN + + P KQLLSFL ++
Sbjct: 399 YNPLGLNICCKCFPSKNTNPRNKQLLSFLSTQ 304
>AW687968 similar to GP|22136982|gb| unknown protein {Arabidopsis thaliana},
partial (9%)
Length = 540
Score = 27.7 bits (60), Expect = 8.0
Identities = 11/25 (44%), Positives = 14/25 (56%)
Frame = +2
Query: 340 KAAAVTVSFISSRSRYSPPPPPPID 364
K T+SFI PPPPPP++
Sbjct: 362 KLILTTLSFIHRLFHSDPPPPPPLE 436
>TC92637 similar to PIR|T39903|T39903 serine-rich protein - fission yeast
(Schizosaccharomyces pombe), partial (7%)
Length = 671
Score = 27.7 bits (60), Expect = 8.0
Identities = 10/21 (47%), Positives = 14/21 (66%)
Frame = -1
Query: 354 RYSPPPPPPIDFVADHPFLFL 374
R+ PPPPPP+ + FLF+
Sbjct: 425 RFFPPPPPPLFSSSSSAFLFI 363
>TC76690 similar to GP|21593806|gb|AAM65773.1 putative RING zinc finger
protein {Arabidopsis thaliana}, partial (22%)
Length = 1303
Score = 27.7 bits (60), Expect = 8.0
Identities = 14/40 (35%), Positives = 19/40 (47%)
Frame = -1
Query: 348 FISSRSRYSPPPPPPIDFVADHPFLFLIREEFSGTILFVG 387
F +S PPPP P PF F++R + IL +G
Sbjct: 868 FEASAIELHPPPPQPSHDGETFPFQFILRILLNEDILLIG 749
>TC92049 similar to PIR|T01061|T01061 hypothetical protein YUP8H12R.45 -
Arabidopsis thaliana, partial (16%)
Length = 1079
Score = 23.1 bits (48), Expect(2) = 8.5
Identities = 7/19 (36%), Positives = 14/19 (72%)
Frame = -2
Query: 344 VTVSFISSRSRYSPPPPPP 362
+++S ++ + + PPPPPP
Sbjct: 115 LSLSIATAINPFPPPPPPP 59
Score = 22.7 bits (47), Expect(2) = 8.5
Identities = 12/30 (40%), Positives = 14/30 (46%)
Frame = -2
Query: 357 PPPPPPIDFVADHPFLFLIREEFSGTILFV 386
PPPPPP PFL FS + F+
Sbjct: 79 PPPPPP-------PFLVPFHISFSFSFSFL 11
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.136 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,007,797
Number of Sequences: 36976
Number of extensions: 153680
Number of successful extensions: 3203
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 1646
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2939
length of query: 395
length of database: 9,014,727
effective HSP length: 98
effective length of query: 297
effective length of database: 5,391,079
effective search space: 1601150463
effective search space used: 1601150463
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC122166.2


