

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122166.12 - phase: 0 /pseudo
(170 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
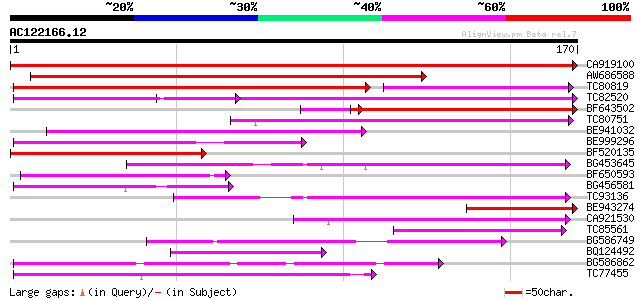
Score E
Sequences producing significant alignments: (bits) Value
CA919100 homologue to PIR|G90291|G902 endoglucanase precursor [i... 339 4e-94
AW686588 153 3e-38
TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non... 110 2e-36
TC82520 105 1e-23
BF643502 100 3e-22
TC80751 weakly similar to GP|13161403|dbj|BAB33036. CPRD49 {Vign... 77 3e-15
BE941032 75 1e-14
BE999296 66 8e-12
BF520135 61 2e-10
BG453645 57 4e-09
BF650593 53 5e-08
BG456581 52 2e-07
TC93136 50 6e-07
BE943274 49 7e-07
CA921530 48 2e-06
TC85561 weakly similar to GP|4530126|gb|AAD21872.1| receptor-lik... 47 3e-06
BG586749 weakly similar to GP|18138053|emb tropinone reductase I... 47 5e-06
BQ124492 weakly similar to GP|9909168|dbj| putative transposable... 46 8e-06
BG586862 45 1e-05
TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarot... 44 2e-05
>CA919100 homologue to PIR|G90291|G902 endoglucanase precursor [imported] -
Sulfolobus solfataricus, partial (3%)
Length = 789
Score = 339 bits (869), Expect = 4e-94
Identities = 159/170 (93%), Positives = 163/170 (95%)
Frame = -2
Query: 1 MIWHIQVPLKVSVFAWRLLRDRLPTKSNLVYRGVIPSEAGLCVSG*GVLESAQHLFLSCS 60
MIWH QVPLKVSVFAWRLLRDRLPTKSNL+YRGVIP+EAGLCVSG G LESAQHLFLSCS
Sbjct: 587 MIWHRQVPLKVSVFAWRLLRDRLPTKSNLIYRGVIPTEAGLCVSGCGALESAQHLFLSCS 408
Query: 61 YFALLWSLVRDWIGFVGVDTNVLSDHFVQFVHSTGGSKASQSFLQLIWLLCAWVLWTERN 120
YFA LWSLVRDWIGFVGVDTNVLSDHFVQFVHSTGG+KASQSFLQLIWLLCAWVLWTERN
Sbjct: 407 YFASLWSLVRDWIGFVGVDTNVLSDHFVQFVHSTGGNKASQSFLQLIWLLCAWVLWTERN 228
Query: 121 NMCFNDSIIPIPRLLDKVKYLSLGWLKTRKASFLFGTFSWWSNPLQCLGI 170
NMCFNDSI P+PRLLDKVKYLSLGWLK R ASFLFGTFSWWSNPLQCLGI
Sbjct: 227 NMCFNDSITPLPRLLDKVKYLSLGWLKARNASFLFGTFSWWSNPLQCLGI 78
>AW686588
Length = 567
Score = 153 bits (387), Expect = 3e-38
Identities = 69/119 (57%), Positives = 87/119 (72%)
Frame = +1
Query: 7 VPLKVSVFAWRLLRDRLPTKSNLVYRGVIPSEAGLCVSG*GVLESAQHLFLSCSYFALLW 66
VPLKVS+ AWRL+RDRLPTK+NLV R + EA CV G G+ E+A HLFL C+ F +W
Sbjct: 142 VPLKVSILAWRLIRDRLPTKANLVRRRCLAVEAAGCVVGCGIAETANHLFLHCATFGAVW 321
Query: 67 SLVRDWIGFVGVDTNVLSDHFVQFVHSTGGSKASQSFLQLIWLLCAWVLWTERNNMCFN 125
+R WIG G D + LSDHF+QF+ TG ++A +SF+QLIWLLC W++W ERNN FN
Sbjct: 322 QHIRAWIGVSGADPHDLSDHFIQFITCTGHTRARRSFMQLIWLLCVWMVWNERNNRLFN 498
>TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non-LTR
retroelement reverse transcriptase {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 1262
Score = 110 bits (275), Expect(2) = 2e-36
Identities = 51/107 (47%), Positives = 68/107 (62%)
Frame = +2
Query: 2 IWHIQVPLKVSVFAWRLLRDRLPTKSNLVYRGVIPSEAGLCVSG*GVLESAQHLFLSCSY 61
+WH +P KVS+F WRLLR+RLPTK NLV+RGV+ + CV G ES HLFL C+
Sbjct: 626 VWHKHIPSKVSLFVWRLLRNRLPTKDNLVHRGVLLATNAACVCGCVDSESTTHLFLHCNV 805
Query: 62 FALLWSLVRDWIGFVGVDTNVLSDHFVQFVHSTGGSKASQSFLQLIW 108
F LWSLVR+W+G + + L HF+QF G + S + +LI+
Sbjct: 806 FCSLWSLVRNWLGIPSMSSGELRTHFIQFAKMVGMPRVSHLYFRLIF 946
Score = 58.5 bits (140), Expect(2) = 2e-36
Identities = 22/57 (38%), Positives = 34/57 (59%)
Frame = +1
Query: 113 WVLWTERNNMCFNDSIIPIPRLLDKVKYLSLGWLKTRKASFLFGTFSWWSNPLQCLG 169
WV+W ERNN F +++ L+D++K S WLK+++ F + WW N L C+G
Sbjct: 946 WVIWKERNNRLFQNTVTAPFVLIDRIKLHSFLWLKSKQVGFAYSYLDWWKNSLLCMG 1116
>TC82520
Length = 833
Score = 105 bits (261), Expect = 1e-23
Identities = 49/126 (38%), Positives = 70/126 (54%)
Frame = +2
Query: 45 G*GVLESAQHLFLSCSYFALLWSLVRDWIGFVGVDTNVLSDHFVQFVHSTGGSKASQSFL 104
G G E+A HLFL C F LWS V W+ + V + F+QF G + + SFL
Sbjct: 314 GCGDSETATHLFLHCDIFGSLWSHVLRWLHLLLVLPADIRQFFIQFTSMAGSPRFTHSFL 493
Query: 105 QLIWLLCAWVLWTERNNMCFNDSIIPIPRLLDKVKYLSLGWLKTRKASFLFGTFSWWSNP 164
Q++W WVLW +RNN F +S+ +++VK S WLK ++A+F F WW +P
Sbjct: 494 QIMWFASVWVLWKKRNNRVFQNSLSDPSTFVEQVKMHSFLWLKFQQATFSFSYHDWWKHP 673
Query: 165 LQCLGI 170
L C+G+
Sbjct: 674 LLCMGV 691
Score = 52.8 bits (125), Expect = 7e-08
Identities = 26/68 (38%), Positives = 37/68 (54%)
Frame = +3
Query: 2 IWHIQVPLKVSVFAWRLLRDRLPTKSNLVYRGVIPSEAGLCVSG*GVLESAQHLFLSCSY 61
+W +P KVS+F WRL +RLPTK NL+ R V+ C+SG + QH+
Sbjct: 186 VWQKNIPSKVSMFVWRLFHNRLPTKVNLMQRHVLQQTHTACISG-VAIRKRQHICFYIVI 362
Query: 62 FALLWSLV 69
F L+ L+
Sbjct: 363 FLALFGLM 386
>BF643502
Length = 631
Score = 99.8 bits (247), Expect(2) = 3e-22
Identities = 44/68 (64%), Positives = 52/68 (75%)
Frame = -1
Query: 103 FLQLIWLLCAWVLWTERNNMCFNDSIIPIPRLLDKVKYLSLGWLKTRKASFLFGTFSWWS 162
F+QLIWLLC WV+W ERNN+ FN + PIP LLDK+K LSLGWLK + A+F+FG S
Sbjct: 502 FMQLIWLLCVWVVWNERNNVMFNHVVTPIPCLLDKIKLLSLGWLKAKNATFVFGIQQGRS 323
Query: 163 NPLQCLGI 170
NPL CLGI
Sbjct: 322 NPLVCLGI 299
Score = 21.2 bits (43), Expect(2) = 3e-22
Identities = 9/19 (47%), Positives = 10/19 (52%)
Frame = -2
Query: 88 VQFVHSTGGSKASQSFLQL 106
+QF H TGG A S L
Sbjct: 546 LQFTHLTGGGVARHSLCNL 490
>TC80751 weakly similar to GP|13161403|dbj|BAB33036. CPRD49 {Vigna
unguiculata}, partial (52%)
Length = 1156
Score = 77.0 bits (188), Expect = 3e-15
Identities = 40/104 (38%), Positives = 56/104 (53%), Gaps = 1/104 (0%)
Frame = +1
Query: 67 SLVRDW-IGFVGVDTNVLSDHFVQFVHSTGGSKASQSFLQLIWLLCAWVLWTERNNMCFN 125
S + W +G D + DH +Q V G S++ +S L L+W WV+W ER+ M FN
Sbjct: 682 SFIHKWFLGVSSTDPPCIPDHLIQLVQ*GGYSRSIRSTLLLVWFSYMWVIWKERSKMIFN 861
Query: 126 DSIIPIPRLLDKVKYLSLGWLKTRKASFLFGTFSWWSNPLQCLG 169
+ +LL+KVK LS WLK ++F F S NPL C+G
Sbjct: 862 QKKDHVHQLLEKVKLLSF*WLKATNSTFPFDYHSRRLNPLLCMG 993
>BE941032
Length = 435
Score = 75.1 bits (183), Expect = 1e-14
Identities = 41/96 (42%), Positives = 52/96 (53%)
Frame = +2
Query: 12 SVFAWRLLRDRLPTKSNLVYRGVIPSEAGLCVSG*GVLESAQHLFLSCSYFALLWSLVRD 71
S+F W +L +R PTK NL+ RGVI + CV G L A HLFL C++F +W V D
Sbjct: 146 SIFLWCVLLNRFPTKDNLLKRGVISAIYQSCVGECGNLYDATHLFLHCNFFRQIWINVSD 325
Query: 72 WIGFVGVDTNVLSDHFVQFVHSTGGSKASQSFLQLI 107
W+ FV V +SD QF G S +QLI
Sbjct: 326 WLSFVMVTLLRISDRLAQFGSFGGFSNCKWDTIQLI 433
>BE999296
Length = 384
Score = 65.9 bits (159), Expect = 8e-12
Identities = 38/88 (43%), Positives = 47/88 (53%)
Frame = -1
Query: 2 IWHIQVPLKVSVFAWRLLRDRLPTKSNLVYRGVIPSEAGLCVSG*GVLESAQHLFLSCSY 61
+WH +PLKV F R+LR+ LPTK N V R VI E LC +G E+ LF
Sbjct: 240 VWHNHIPLKVCFFVLRVLRNCLPTKDNFVRRRVIHEEHMLCPTGCSFKETTDDLF----- 76
Query: 62 FALLWSLVRDWIGFVGVDTNVLSDHFVQ 89
LW LV W+ V++ VL DHF Q
Sbjct: 75 ---LWPLV*QWLHIS*VNSCVLRDHFYQ 1
>BF520135
Length = 202
Score = 60.8 bits (146), Expect = 2e-10
Identities = 27/59 (45%), Positives = 40/59 (67%)
Frame = +3
Query: 1 MIWHIQVPLKVSVFAWRLLRDRLPTKSNLVYRGVIPSEAGLCVSG*GVLESAQHLFLSC 59
++W + KVS+FAWRL RLPTK+N+ RG++ +A +CV+ ++ES HLFL C
Sbjct: 12 LLWRKEDLSKVSIFAWRLFHGRLPTKANVFKRGIVHHDAHMCVTRCRLIESDVHLFLHC 188
>BG453645
Length = 622
Score = 57.0 bits (136), Expect = 4e-09
Identities = 41/138 (29%), Positives = 62/138 (44%), Gaps = 5/138 (3%)
Frame = +3
Query: 36 PSEAGLCVSG*GVLESAQHLFLSCSYFALLWSLVRDWIGFVGVDTNVLSDHFVQFVH--- 92
P + CV +E+ H+FL C +L+W V W D N L + FVH
Sbjct: 15 PEVSSTCVLCNLKVETTTHIFLHCDVASLVWFRVMKWF-----DVNFLIPPNL-FVHWEC 176
Query: 93 -STGGSKASQSFLQ-LIWLLCAWVLWTERNNMCFNDSIIPIPRLLDKVKYLSLGWLKTRK 150
S GGS + L+W WVLW +RN++ F +++++K LS W+ R
Sbjct: 177 WSEGGSANRVTKGHWLVWHTTIWVLWAKRNDLIFKGLNCVAEDVIEEIKVLS*RWMLERS 356
Query: 151 ASFLFGTFSWWSNPLQCL 168
++ + W NP CL
Sbjct: 357 STPSCFFYEWSWNPRLCL 410
>BF650593
Length = 486
Score = 53.1 bits (126), Expect = 5e-08
Identities = 27/63 (42%), Positives = 34/63 (53%)
Frame = +3
Query: 4 HIQVPLKVSVFAWRLLRDRLPTKSNLVYRGVIPSEAGLCVSG*GVLESAQHLFLSCSYFA 63
H VPLKVS WRL ++ L T+ NL RGV+ + CV G ES H F C F+
Sbjct: 297 HKSVPLKVSCLVWRLFQNXLATRDNLSKRGVLDQNSIXCVXDCGREESVSHFFFECP-FS 473
Query: 64 LLW 66
+W
Sbjct: 474 XVW 482
>BG456581
Length = 683
Score = 51.6 bits (122), Expect = 2e-07
Identities = 27/67 (40%), Positives = 35/67 (51%), Gaps = 1/67 (1%)
Frame = +2
Query: 2 IWHIQVPLKVSVFAWRLLRDRLPTKSNLVYRG-VIPSEAGLCVSG*GVLESAQHLFLSCS 60
IW+ +P S WRL DRLPT NL RG + S C+ +E++ HLFL C
Sbjct: 80 IWNSCIPPSHSFICWRLAHDRLPTDDNLSSRGCALVSMCSFCLE---QVETSDHLFLRCK 250
Query: 61 YFALLWS 67
+ LWS
Sbjct: 251 FVVTLWS 271
>TC93136
Length = 722
Score = 49.7 bits (117), Expect = 6e-07
Identities = 31/119 (26%), Positives = 52/119 (43%)
Frame = +1
Query: 50 ESAQHLFLSCSYFALLWSLVRDWIGFVGVDTNVLSDHFVQFVHSTGGSKASQSFLQLIWL 109
E++ HLFL C + + +WS + W+ + FV V ++ + LIW
Sbjct: 262 ETSSHLFLHCPFLSSVWSKILGWLYY---------SKFV-CVFRLLERRSRTKGVWLIWH 411
Query: 110 LCAWVLWTERNNMCFNDSIIPIPRLLDKVKYLSLGWLKTRKASFLFGTFSWWSNPLQCL 168
WV+W NN F + I +++++K LS W TR + W +P C+
Sbjct: 412 ATIWVIWKGINNRIFKNISKAIDEIVEEIKVLSWRWSLTRLKCPPCFYYEWCWDPGDCV 588
>BE943274
Length = 597
Score = 49.3 bits (116), Expect = 7e-07
Identities = 20/33 (60%), Positives = 23/33 (69%)
Frame = -3
Query: 138 VKYLSLGWLKTRKASFLFGTFSWWSNPLQCLGI 170
V Y SL WLK + +F+FG WWSNPL CLGI
Sbjct: 343 VTYYSLWWLKVKHVTFVFGIQIWWSNPLMCLGI 245
>CA921530
Length = 793
Score = 47.8 bits (112), Expect = 2e-06
Identities = 27/88 (30%), Positives = 40/88 (44%), Gaps = 5/88 (5%)
Frame = -1
Query: 86 HFVQFVHST-----GGSKASQSFLQLIWLLCAWVLWTERNNMCFNDSIIPIPRLLDKVKY 140
HF+ F+ S +K + L LIW W+LW NN FN+ + ++++VK
Sbjct: 670 HFIMFIMSQCWNGWERNKKIRRGLWLIWHATIWILWKAWNNRIFNNQAVEAEDIIEEVKV 491
Query: 141 LSLGWLKTRKASFLFGTFSWWSNPLQCL 168
LS W R + + W NP CL
Sbjct: 490 LSWRWTFNRINIPVCMYYEWCWNPKYCL 407
>TC85561 weakly similar to GP|4530126|gb|AAD21872.1| receptor-like protein
kinase homolog RK20-1 {Phaseolus vulgaris}, partial (18%)
Length = 1549
Score = 47.4 bits (111), Expect = 3e-06
Identities = 21/52 (40%), Positives = 28/52 (53%)
Frame = +2
Query: 116 WTERNNMCFNDSIIPIPRLLDKVKYLSLGWLKTRKASFLFGTFSWWSNPLQC 167
W ERN+ FND++ +LL +K+ S WLK K F F + WWS C
Sbjct: 1259 WKERNSRIFNDNMSSKDQLLASIKFHS*WWLKMHKLGFAFDYYYWWSMASLC 1414
>BG586749 weakly similar to GP|18138053|emb tropinone reductase I {Calystegia
sepium}, partial (11%)
Length = 787
Score = 46.6 bits (109), Expect = 5e-06
Identities = 30/108 (27%), Positives = 53/108 (48%)
Frame = -2
Query: 42 CVSG*GVLESAQHLFLSCSYFALLWSLVRDWIGFVGVDTNVLSDHFVQFVHSTGGSKASQ 101
CV GV E ++HLFL C+ A +W+ + W+GF L H+ + +G K +
Sbjct: 531 CVLCEGVPEVSEHLFLQCAT-AKVWADLLRWLGFDYHTPPDLFFHWKWWNEMSGNKKTRK 355
Query: 102 SFLQLIWLLCAWVLWTERNNMCFNDSIIPIPRLLDKVKYLSLGWLKTR 149
+ ++W RN++ FN+S + L++ +K L+ W+ R
Sbjct: 354 GY*---------IIWRVRNDLIFNNSRSGMDELVEAIKVLAWRWVLNR 238
>BQ124492 weakly similar to GP|9909168|dbj| putative transposable element
Tip100 protein {Oryza sativa (japonica cultivar-group)},
partial (8%)
Length = 694
Score = 45.8 bits (107), Expect = 8e-06
Identities = 20/47 (42%), Positives = 27/47 (56%)
Frame = +2
Query: 49 LESAQHLFLSCSYFALLWSLVRDWIGFVGVDTNVLSDHFVQFVHSTG 95
+++A+HLFL C F+ LWS V W+G V L HF QF + G
Sbjct: 527 IKTARHLFLDCDIFSSLWSQVWLWLGISSVQPGELRHHFPQFTNMAG 667
>BG586862
Length = 804
Score = 45.1 bits (105), Expect = 1e-05
Identities = 37/129 (28%), Positives = 57/129 (43%)
Frame = -1
Query: 2 IWHIQVPLKVSVFAWRLLRDRLPTKSNLVYRGVIPSEAGLCVSG*GVLESAQHLFLSCSY 61
+W I+ + F WRLL + LP K L RG+ S LC +E+ QHLFL+C
Sbjct: 654 VWGIKTIPRHKSFLWRLLHNALPVKDELHKRGIRCSL--LCPRCESKIETVQHLFLNCEV 481
Query: 62 FALLWSLVRDWIGFVGVDTNVLSDHFVQFVHSTGGSKASQSFLQLIWLLCAWVLWTERNN 121
W +G + VL HF ++ + ++ + L LL + +W RN
Sbjct: 480 TQKEW--FGSQLGINFHSSGVL--HFHDWITNFILKNDEETIIALTALL--YSIWHARNQ 319
Query: 122 MCFNDSIIP 130
F + +P
Sbjct: 318 KVFENIDVP 292
>TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarotenoid
dioxygenase1 {Pisum sativum}, partial (43%)
Length = 1865
Score = 44.3 bits (103), Expect = 2e-05
Identities = 32/110 (29%), Positives = 47/110 (42%), Gaps = 1/110 (0%)
Frame = -2
Query: 2 IWHIQVPLKVSVFAWRLLRDRLPTKSNLVYRGVIPSE-AGLCVSG*GVLESAQHLFLSCS 60
+W + P KV F+W L DR+PT NL R ++ E + CV E+ HLFL C
Sbjct: 322 LWKSKAPAKVLAFSWTLFLDRIPTMVNLGKRRLLRVEDSKRCVFCGCQDETVVHLFLHCD 143
Query: 61 YFALLWSLVRDWIGFVGVDTNVLSDHFVQFVHSTGGSKASQSFLQLIWLL 110
+ + V W+ F + L H + + K Q WL+
Sbjct: 142 VISKV*REVMRWLNFNLISPPNLLIHAICWSREVRAKKLRQG----AWLI 5
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.331 0.143 0.497
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,924,314
Number of Sequences: 36976
Number of extensions: 136585
Number of successful extensions: 985
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 968
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 976
length of query: 170
length of database: 9,014,727
effective HSP length: 89
effective length of query: 81
effective length of database: 5,723,863
effective search space: 463632903
effective search space used: 463632903
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 55 (25.8 bits)
Medicago: description of AC122166.12


