

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122166.11 + phase: 0
(554 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
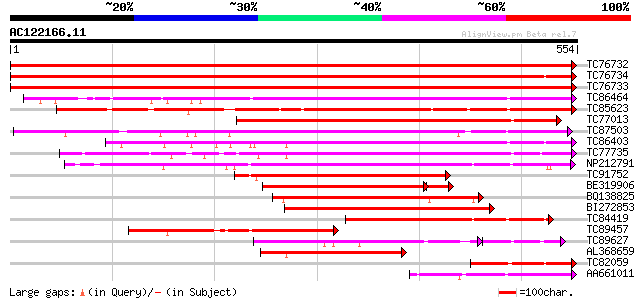
Score E
Sequences producing significant alignments: (bits) Value
TC76732 homologue to PIR|T06468|T06468 pectinesterase (EC 3.1.1.... 1098 0.0
TC76734 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11... 951 0.0
TC76733 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11... 927 0.0
TC86464 similar to GP|2098711|gb|AAB57670.1| pectinesterase {Cit... 441 e-124
TC85623 similar to GP|13605696|gb|AAK32841.1 At2g45220/F4L23.27 ... 413 e-116
TC77013 similar to GP|7025485|gb|AAF35897.1| pectin methylestera... 389 e-108
TC87503 similar to SP|Q43111|PME3_PHAVU Pectinesterase 3 precurs... 384 e-107
TC86403 similar to PIR|B86158|B86158 hypothetical protein AAF028... 360 e-100
TC77735 similar to GP|14334646|gb|AAK59501.1 putative pectineste... 339 2e-93
NP212791 NP212791|AJ249611.1|CAB65291.1 pectin methyl-esterase PEF1 291 4e-79
TC91752 similar to GP|6630559|gb|AAF19578.1| putative pectineste... 260 8e-70
BE319906 similar to SP|Q43062|PME_ Pectinesterase PPE8B precurso... 204 3e-57
BQ138825 similar to PIR|T49922|T49 pectin methylesterase-like pr... 203 1e-52
BI272853 weakly similar to PIR|T04359|T04 pectin methylesterase-... 190 1e-48
TC84419 similar to SP|Q42920|PME_MEDSA Pectinesterase precursor ... 184 1e-46
TC89457 weakly similar to SP|Q43062|PME_PRUPE Pectinesterase PPE... 159 3e-39
TC89627 weakly similar to GP|17529290|gb|AAL38872.1 putative pec... 119 3e-38
AL368659 similar to PIR|B86158|B86 hypothetical protein AAF02886... 149 4e-36
TC82059 similar to GP|19424045|gb|AAL87311.1 unknown protein {Ar... 126 2e-29
AA661011 weakly similar to PIR|T10494|T104 pectinesterase (EC 3.... 122 3e-28
>TC76732 homologue to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11)
precursor - garden pea, complete
Length = 1860
Score = 1098 bits (2839), Expect = 0.0
Identities = 554/554 (100%), Positives = 554/554 (100%)
Frame = +2
Query: 1 MAIQQSLLDKPRKSIPKTFWLVLSLVAIISSSALIISHLNKPISFFNLSSAPNLCEHALD 60
MAIQQSLLDKPRKSIPKTFWLVLSLVAIISSSALIISHLNKPISFFNLSSAPNLCEHALD
Sbjct: 35 MAIQQSLLDKPRKSIPKTFWLVLSLVAIISSSALIISHLNKPISFFNLSSAPNLCEHALD 214
Query: 61 TKSCLTHVSEVVQGSTLSNTKDHKLSTLVSLLTKSTAHIRKAMDTANVIKRRVNSPREEI 120
TKSCLTHVSEVVQGSTLSNTKDHKLSTLVSLLTKSTAHIRKAMDTANVIKRRVNSPREEI
Sbjct: 215 TKSCLTHVSEVVQGSTLSNTKDHKLSTLVSLLTKSTAHIRKAMDTANVIKRRVNSPREEI 394
Query: 121 ALNDCEELMDLSMDRVWDSVLTLTKNNIDSQHDAHTWLSSVLTNHATCLDGLEGSSRVVM 180
ALNDCEELMDLSMDRVWDSVLTLTKNNIDSQHDAHTWLSSVLTNHATCLDGLEGSSRVVM
Sbjct: 395 ALNDCEELMDLSMDRVWDSVLTLTKNNIDSQHDAHTWLSSVLTNHATCLDGLEGSSRVVM 574
Query: 181 ESDLHDLISRARSSLAVLVSVLPPKANDGFIDEKLNGDFPSWVTSKDRRLLESSVGDIKA 240
ESDLHDLISRARSSLAVLVSVLPPKANDGFIDEKLNGDFPSWVTSKDRRLLESSVGDIKA
Sbjct: 575 ESDLHDLISRARSSLAVLVSVLPPKANDGFIDEKLNGDFPSWVTSKDRRLLESSVGDIKA 754
Query: 241 NVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKGTYKENIEIGKKKTNVMLVGDGMD 300
NVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKGTYKENIEIGKKKTNVMLVGDGMD
Sbjct: 755 NVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKGTYKENIEIGKKKTNVMLVGDGMD 934
Query: 301 ATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRVGADQSVIN 360
ATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRVGADQSVIN
Sbjct: 935 ATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRVGADQSVIN 1114
Query: 361 RCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKPMANQKNMVTA 420
RCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKPMANQKNMVTA
Sbjct: 1115RCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKPMANQKNMVTA 1294
Query: 421 QGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTVVLQSVVDGHIDPA 480
QGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTVVLQSVVDGHIDPA
Sbjct: 1295QGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTVVLQSVVDGHIDPA 1474
Query: 481 GWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASKFTVTQLIQGN 540
GWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASKFTVTQLIQGN
Sbjct: 1475GWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASKFTVTQLIQGN 1654
Query: 541 VWLKNTGVAFIEGL 554
VWLKNTGVAFIEGL
Sbjct: 1655VWLKNTGVAFIEGL 1696
>TC76734 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11) precursor
- garden pea, complete
Length = 1981
Score = 951 bits (2457), Expect = 0.0
Identities = 471/554 (85%), Positives = 512/554 (92%)
Frame = +3
Query: 1 MAIQQSLLDKPRKSIPKTFWLVLSLVAIISSSALIISHLNKPISFFNLSSAPNLCEHALD 60
MA Q++LL KPRKSIPKTFWLVLSL AIISSSALIISHLNKPIS F+ SSAPN+CEHA+D
Sbjct: 42 MATQETLLHKPRKSIPKTFWLVLSLAAIISSSALIISHLNKPISIFHFSSAPNVCEHAVD 221
Query: 61 TKSCLTHVSEVVQGSTLSNTKDHKLSTLVSLLTKSTAHIRKAMDTANVIKRRVNSPREEI 120
T SCLTHV+EVVQGSTL NTKDHKLSTL+SLLTKST HIRKAMDTANVIKRR+NSPREE
Sbjct: 222 TNSCLTHVAEVVQGSTLDNTKDHKLSTLISLLTKSTTHIRKAMDTANVIKRRINSPREEN 401
Query: 121 ALNDCEELMDLSMDRVWDSVLTLTKNNIDSQHDAHTWLSSVLTNHATCLDGLEGSSRVVM 180
ALN CE+LM+LSM+RVWDSVLTLTK+N+DSQ DAHTWLSSVLTNHATCLDGLEG+SR VM
Sbjct: 402 ALNVCEKLMNLSMERVWDSVLTLTKDNMDSQQDAHTWLSSVLTNHATCLDGLEGTSRAVM 581
Query: 181 ESDLHDLISRARSSLAVLVSVLPPKANDGFIDEKLNGDFPSWVTSKDRRLLESSVGDIKA 240
E+D+ DLI+RARSSLAVLV+VLPPK +D FIDE LNGDFPSWVTSKDRRLLESSVGD+KA
Sbjct: 582 ENDIQDLIARARSSLAVLVAVLPPKDHDEFIDESLNGDFPSWVTSKDRRLLESSVGDVKA 761
Query: 241 NVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKGTYKENIEIGKKKTNVMLVGDGMD 300
NVVVA+DGSGKFKTVA+AVASAP+ G RYVIYVKKG YKEN+EI KTNVML+GDGMD
Sbjct: 762 NVVVAKDGSGKFKTVAEAVASAPNKGTARYVIYVKKGIYKENVEIASSKTNVMLLGDGMD 941
Query: 301 ATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRVGADQSVIN 360
ATIITGSLN++DGT TF++ATVAAVGD FIAQDI FQNTAGPQKHQAVALRVG+D+SVIN
Sbjct: 942 ATIITGSLNYVDGTGTFQTATVAAVGDWFIAQDIGFQNTAGPQKHQAVALRVGSDRSVIN 1121
Query: 361 RCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKPMANQKNMVTA 420
RCKIDAFQDTLYAH+NRQFYRDS+ITGT+DFIFG+AAVV QK KL ARKPMANQ NMVTA
Sbjct: 1122RCKIDAFQDTLYAHTNRQFYRDSFITGTIDFIFGDAAVVLQKCKLVARKPMANQNNMVTA 1301
Query: 421 QGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTVVLQSVVDGHIDPA 480
QGR DPNQNTATSIQQCDVIPS+DLKPV GS+KTYLGRPWKKYSRTVV+QS++ HIDP
Sbjct: 1302QGRIDPNQNTATSIQQCDVIPSTDLKPVIGSVKTYLGRPWKKYSRTVVMQSLLGAHIDPT 1481
Query: 481 GWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASKFTVTQLIQGN 540
GWAEWDAASKDFLQTLYYGEYMNSG GAGT KRV WPGYHII N AEA+KFTV QLIQGN
Sbjct: 1482GWAEWDAASKDFLQTLYYGEYMNSGPGAGTSKRVKWPGYHII-NTAEANKFTVAQLIQGN 1658
Query: 541 VWLKNTGVAFIEGL 554
VWLKNTGVAFI GL
Sbjct: 1659VWLKNTGVAFIAGL 1700
>TC76733 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11) precursor
- garden pea, partial (98%)
Length = 2029
Score = 927 bits (2397), Expect = 0.0
Identities = 462/555 (83%), Positives = 500/555 (89%), Gaps = 1/555 (0%)
Frame = +2
Query: 1 MAIQQSLLDKPRKSIPKTFWLVLSLVAIISSSALIISHLNKPISFFNLSSAPNL-CEHAL 59
MA QQSLLDKPRKS+PKTFWL+LSLVAII SSALI +HL KPISFF+L++ N+ CEHA+
Sbjct: 47 MANQQSLLDKPRKSLPKTFWLILSLVAIIISSALISTHLKKPISFFHLTTVQNVYCEHAV 226
Query: 60 DTKSCLTHVSEVVQGSTLSNTKDHKLSTLVSLLTKSTAHIRKAMDTANVIKRRVNSPREE 119
DTKSCL HVSEV TL TKD L L+SLLTKST HI+ AMDTA+VIKRR+NSPREE
Sbjct: 227 DTKSCLAHVSEVSHVPTLVTTKDQNLHVLLSLLTKSTTHIQNAMDTASVIKRRINSPREE 406
Query: 120 IALNDCEELMDLSMDRVWDSVLTLTKNNIDSQHDAHTWLSSVLTNHATCLDGLEGSSRVV 179
IAL+DCE+LMDLSM+R+WD++L LTKNNIDSQ DAHTWLSSVLTNHATCLDGLEGSSRVV
Sbjct: 407 IALSDCEQLMDLSMNRIWDTMLKLTKNNIDSQQDAHTWLSSVLTNHATCLDGLEGSSRVV 586
Query: 180 MESDLHDLISRARSSLAVLVSVLPPKANDGFIDEKLNGDFPSWVTSKDRRLLESSVGDIK 239
ME+DL DLISRARSSLAV + V P K D FIDE L G+FPSWVTSKDRRLLE++VGDIK
Sbjct: 587 MENDLQDLISRARSSLAVFLVVFPQKDRDQFIDETLIGEFPSWVTSKDRRLLETAVGDIK 766
Query: 240 ANVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKGTYKENIEIGKKKTNVMLVGDGM 299
ANVVVAQDGSGKFKTVA+AVASAPDNGKT+YVIYVKKGTYKEN+EIG KKTNVMLVGDGM
Sbjct: 767 ANVVVAQDGSGKFKTVAEAVASAPDNGKTKYVIYVKKGTYKENVEIGSKKTNVMLVGDGM 946
Query: 300 DATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRVGADQSVI 359
DATIITG+LNFIDGTTTFKS+TVAAVGDGFIAQDI FQN AG KHQAVALRVG+DQSVI
Sbjct: 947 DATIITGNLNFIDGTTTFKSSTVAAVGDGFIAQDIWFQNMAGAAKHQAVALRVGSDQSVI 1126
Query: 360 NRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKPMANQKNMVT 419
NRC+IDAFQDTLYAHSNRQFYRDS ITGT+DFIFGNAAVVFQK KL ARKPMANQ NM T
Sbjct: 1127NRCRIDAFQDTLYAHSNRQFYRDSVITGTIDFIFGNAAVVFQKCKLVARKPMANQNNMFT 1306
Query: 420 AQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTVVLQSVVDGHIDP 479
AQGREDP QNT TSIQQCD+ PSSDLKPV GSIKT+LGRPWKKYSRTVV+QS +D HIDP
Sbjct: 1307AQGREDPGQNTGTSIQQCDLTPSSDLKPVVGSIKTFLGRPWKKYSRTVVMQSFLDSHIDP 1486
Query: 480 AGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASKFTVTQLIQG 539
GWAEWDAASKDFLQTLYYGEY+N+G GAGT KRVTWPGYH+I AAEASKFTV QLIQG
Sbjct: 1487TGWAEWDAASKDFLQTLYYGEYLNNGPGAGTAKRVTWPGYHVINTAAEASKFTVAQLIQG 1666
Query: 540 NVWLKNTGVAFIEGL 554
NVWLKNTGVAF EGL
Sbjct: 1667NVWLKNTGVAFTEGL 1711
>TC86464 similar to GP|2098711|gb|AAB57670.1| pectinesterase {Citrus
sinensis}, partial (85%)
Length = 1993
Score = 441 bits (1133), Expect = e-124
Identities = 261/583 (44%), Positives = 350/583 (59%), Gaps = 42/583 (7%)
Frame = +1
Query: 14 SIPKTFWLVLSLVAI---------------ISSSALIISHLNKPI--SFFNLSSAPNLCE 56
S+ T +V S+VAI I+SS+L +SH + I S + P LC
Sbjct: 115 SLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHAILKSACTTTLYPELCF 294
Query: 57 HALDTKSCLTHVSEVVQGSTLSNTKDHKLSTLVSLLTKSTAHIRKAMDTANVIKRRVNSP 116
A+ ++ +TH ++N KD +S +++ T++ H ++ R+ +
Sbjct: 295 SAISSEPNITH--------KITNHKD-VISLSLNITTRAVEH--NYFTVEKLLLRKSLTK 441
Query: 117 REEIALNDCEELMDLSMDRVW----DSVLTLTKNNIDSQH--DAHTWLSSVLTNHATCLD 170
RE+IAL+DC E +D ++D + D VL +K + QH D T +SS +TN TCLD
Sbjct: 442 REKIALHDCLETIDETLDELKEAQNDLVLYPSKKTL-YQHADDLKTLISSAITNQVTCLD 618
Query: 171 GLEGSS------RVVMESDLH-------------DLISRARSSLAVLVSVLPPKANDGFI 211
G +V+ E +H ++ + + VL N +
Sbjct: 619 GFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKLL 798
Query: 212 DEKLNGDFPSWVTSKDRRLLESSVGDIKANVVVAQDGSGKFKTVAQAVASAPDNGKTRYV 271
+E+ +P W+++ DRRLL+ S +KA+VVVA DGSG FKTV++AVA+AP RYV
Sbjct: 799 EEENGVGWPEWISAGDRRLLQGST--VKADVVVAADGSGNFKTVSEAVAAAPLKSSKRYV 972
Query: 272 IYVKKGTYKENIEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIA 331
I +K G YKEN+E+ KKKTN+M +GDG TIITGS N +DG+TTF SATVA VG F+A
Sbjct: 973 IKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLA 1152
Query: 332 QDIRFQNTAGPQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDF 391
+DI FQNTAGP KHQAVALRVGAD S C I A+QDTLY H+NRQF+ + +I+GTVDF
Sbjct: 1153RDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDF 1332
Query: 392 IFGNAAVVFQKSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGS 451
IFGN+AVVFQ + AR+P + QKNMVTAQGR DPNQNT IQ+C + + DL+ V+G+
Sbjct: 1333IFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGN 1512
Query: 452 IKTYLGRPWKKYSRTVVLQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTG 511
TYLGRPWK+YSRTV +QS + IDP GW EW+ L TL Y EY N+G GAGT
Sbjct: 1513FPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWN--GNFALNTLVYREYQNTGPGAGTS 1686
Query: 512 KRVTWPGYHIIKNAAEASKFTVTQLIQGNVWLKNTGVAFIEGL 554
KRVTW G+ +I +AAEA T I G+ WL +TG F GL
Sbjct: 1687KRVTWKGFKVITSAAEAQSSTPGNFIGGSSWLGSTGFPFSLGL 1815
>TC85623 similar to GP|13605696|gb|AAK32841.1 At2g45220/F4L23.27 {Arabidopsis
thaliana}, partial (79%)
Length = 2442
Score = 413 bits (1062), Expect = e-116
Identities = 237/512 (46%), Positives = 313/512 (60%), Gaps = 3/512 (0%)
Frame = +3
Query: 46 FNLSSAPNLCEHALDTKSCLTHVSEVVQGSTLSNTKDHKLSTLVSLLTKSTAHIRKAMDT 105
++LSS + C + C +++ T+ + D +L + ++ +K +
Sbjct: 636 YSLSSIKSWCSQTPYPQPCEYYLTNNAFNQTIKSKSDFFKVSLEIAMERA----KKGEEN 803
Query: 106 ANVIKRRVNSPREEIALNDCEELMDLSMDRVWDSVLTLTKNNIDSQHDAHTWLSSVLTNH 165
+ + + SP+E+ A DC EL D ++ + L+ TK +Q+++ TWLS+ LTN
Sbjct: 804 THAVGPKCRSPQEKAAWADCLELYDFTVQK-----LSQTKYTKCTQYESQTWLSTALTNL 968
Query: 166 ATCLDGLE--GSSRVVMESDLHDLISRARSSLAV-LVSVLPPKANDGFIDEKLNGDFPSW 222
TC +G G + V+ +++ ++L++ V P DGF P+W
Sbjct: 969 ETCKNGFYDLGVTNYVLPLLSNNVTKLLSNTLSLNKVPYQQPSYKDGF---------PTW 1121
Query: 223 VTSKDRRLLESSVGDIKANVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKGTYKEN 282
V DR+LL++S KANVVVA+DGSGK+ TV A +AP +G RYVIYVK G Y E
Sbjct: 1122 VKPGDRKLLQTSSAASKANVVVAKDGSGKYTTVKAATDAAP-SGSGRYVIYVKAGVYNEQ 1298
Query: 283 IEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGP 342
+EI K NVMLVGDG+ TIITGS + GTTTF+SATVAA GDGFIAQDI F+NTAG
Sbjct: 1299 VEI--KAKNVMLVGDGIGKTIITGSKSVGGGTTTFRSATVAATGDGFIAQDITFRNTAGA 1472
Query: 343 QKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQK 402
HQAVA R G+D SV +C + +QDTLY HS RQFYR+ I GTVDFIFGNAAVV Q
Sbjct: 1473 ANHQAVAFRSGSDLSVFYKCSFEGYQDTLYVHSERQFYRECNIYGTVDFIFGNAAVVLQN 1652
Query: 403 SKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKK 462
+ AR P A + VTAQGR DPNQNT I V SDL P S+K+YLGRPW+K
Sbjct: 1653 CNIFARNPPA-KTITVTAQGRTDPNQNTGIIIHNSRVSAQSDLNP--SSVKSYLGRPWQK 1823
Query: 463 YSRTVVLQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHII 522
YSRTV +++V+DG I+PAGW WD L TLYY EY N+G+G+ T RVTW GYH++
Sbjct: 1824 YSRTVFMKTVLDGFINPAGWLPWD--GNFALDTLYYAEYANTGSGSSTSNRVTWKGYHVL 1997
Query: 523 KNAAEASKFTVTQLIQGNVWLKNTGVAFIEGL 554
+A++AS FTV I GN W+ NTGV F GL
Sbjct: 1998 TSASQASPFTVGNFIAGNSWIGNTGVPFTSGL 2093
>TC77013 similar to GP|7025485|gb|AAF35897.1| pectin methylesterase isoform
alpha {Vigna radiata}, partial (94%)
Length = 1655
Score = 389 bits (998), Expect = e-108
Identities = 188/318 (59%), Positives = 235/318 (73%)
Frame = +2
Query: 222 WVTSKDRRLLESSVGDIKANVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKGTYKE 281
WV+ KDR+LL+++V K ++VVA+DG+G F T+ +A+A+AP++ TR+VI++K G Y E
Sbjct: 41 WVSPKDRKLLQAAVNQTKFDLVVAKDGTGNFATIGEAIAAAPNSSATRFVIHIKAGAYFE 220
Query: 282 NIEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAG 341
N+E+ KKKTN+MLVGDG+ T++ S N +DG TTF+S+T A VGD FIA+ I F+N+AG
Sbjct: 221 NVEVIKKKTNLMLVGDGIGQTVVKASRNVVDGWTTFQSSTFAVVGDKFIAKGITFENSAG 400
Query: 342 PQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQ 401
P KHQAVA+R GAD S +C A+QDTLY HS RQFYR+ + GTVDFIFGNAAVVFQ
Sbjct: 401 PSKHQAVAVRNGADFSAFYQCSFVAYQDTLYVHSLRQFYRECDVYGTVDFIFGNAAVVFQ 580
Query: 402 KSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWK 461
L ARKP QKN+ TAQGREDPNQNT SI C + ++DL PVQ + K+YLGRPWK
Sbjct: 581 NCNLYARKPDPKQKNLFTAQGREDPNQNTGISILNCKIAAAADLIPVQSTFKSYLGRPWK 760
Query: 462 KYSRTVVLQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHI 521
KYSRTV L S +D IDP GW EW+ L TL+YGEY N G G+ T RVTWPGY +
Sbjct: 761 KYSRTVYLNSFIDNLIDPPGWLEWNGTFA--LDTLFYGEYKNRGPGSNTSARVTWPGYKV 934
Query: 522 IKNAAEASKFTVTQLIQG 539
I NA EAS+FTV Q IQG
Sbjct: 935 ITNATEASQFTVRQFIQG 988
>TC87503 similar to SP|Q43111|PME3_PHAVU Pectinesterase 3 precursor (EC
3.1.1.11) (Pectin methylesterase 3) (PE 3)., partial
(93%)
Length = 2088
Score = 384 bits (986), Expect = e-107
Identities = 229/574 (39%), Positives = 327/574 (56%), Gaps = 27/574 (4%)
Frame = +2
Query: 4 QQSLLDKPRKSIPKTFWLVLSLVA-IISSSALIISHLNKPISFFNLSSAPN--------- 53
QQ+ K RK I + LVA II++ A I+ H + S + +S PN
Sbjct: 110 QQAFQKKTRKRITIIIISSIILVAVIIAAVAGILIHKHNTESSSSPNSLPNTELTPATSL 289
Query: 54 --LCEHALDTKSCLTHVSEVVQGSTLSNTKDHKLSTLVSLLTKSTAHIRKAMDTANVIKR 111
+CE SC + +S + +T + KLS V++ S + + + A
Sbjct: 290 KAVCESTQYPNSCFSSISSLPDSNTTDPEQLFKLSLKVAIDELSKLSLTRFSEKAT---- 457
Query: 112 RVNSPREEIALNDCEELMDLSMDRVWDSVLTLTKN----NIDSQHDAHTWLSSVLTNHAT 167
PR + A+ C+ ++ S+DR+ DS+ T+ + D TWLS+ LT+H T
Sbjct: 458 ---EPRVKKAIGVCDNVLADSLDRLNDSMSTIVDGGKMLSPAKIRDVETWLSAALTDHDT 628
Query: 168 CLDGL----EGSSRVVM---ESDLHDLISRARSSLAVLVSVLPPKANDGFIDE--KLNGD 218
CLD + ++R V+ E + + A +SLA++ V+ +N + +L G+
Sbjct: 629 CLDAVGEVNSTAARGVIPEIERIMRNSTEFASNSLAIVSKVIRLLSNFEVSNHHRRLLGE 808
Query: 219 FPSWVTSKDRRLLESSVGDIKANVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKGT 278
FP W+ + +RRLL + V + + VVA+DGSG++KT+ +A+ R+V+YVKKG
Sbjct: 809 FPEWLGTAERRLLATVVNETVPDAVVAKDGSGQYKTIGEALKLVKKKSLQRFVVYVKKGV 988
Query: 279 YKENIEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQN 338
Y ENI++ K NVM+ GDGM T+++GS N+IDGT TF++AT A G GFIA+DI+F N
Sbjct: 989 YVENIDLDKNTWNVMIYGDGMTETVVSGSRNYIDGTPTFETATFAVKGKGFIAKDIQFLN 1168
Query: 339 TAGPQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAV 398
TAG KHQAVA+R G+DQSV RC +QDTLYAHSNRQFYRD ITGT+DFIFGNAA
Sbjct: 1169TAGASKHQAVAMRSGSDQSVFYRCSFVGYQDTLYAHSNRQFYRDCDITGTIDFIFGNAAA 1348
Query: 399 VFQKSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQC--DVIPSSDLKPVQGSIKTYL 456
VFQ K+ R+PM+NQ N +TAQG++DPNQN+ IQ+ +P +L TYL
Sbjct: 1349VFQNCKIMPRQPMSNQFNTITAQGKKDPNQNSGIVIQKSTFTTLPGDNL-----IAPTYL 1513
Query: 457 GRPWKKYSRTVVLQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTW 516
GRPWK +S T++++S + + P GW W A+ + ++ Y EY N+G GA RV W
Sbjct: 1514GRPWKDFSTTIIMKSEIGSFLKPVGWISW-VANVEPPSSILYAEYQNTGPGADVPGRVKW 1690
Query: 517 PGYHIIKNAAEASKFTVTQLIQGNVWLKNTGVAF 550
GY +A KFTV IQG WL + V F
Sbjct: 1691AGYKPALGDEDAIKFTVDSFIQGPEWLPSASVQF 1792
>TC86403 similar to PIR|B86158|B86158 hypothetical protein AAF02886.1
[imported] - Arabidopsis thaliana, partial (63%)
Length = 1929
Score = 360 bits (925), Expect = e-100
Identities = 218/496 (43%), Positives = 284/496 (56%), Gaps = 35/496 (7%)
Frame = +3
Query: 94 KSTAHIRKAMDTAN--VIKRRVNSPREEIALNDCEELMDLSMDRVWDSVLTLTKNNID-- 149
KS + RK D N + +R S AL DC+ L DL+ D + S T+ K
Sbjct: 228 KSLSQSRKFSDLINKYLQRRSTLSTTALRALQDCQSLSDLNFDFLSSSFQTVNKTTKFLP 407
Query: 150 --SQHDAHTWLSSVLTNHATCLDGLEGSS-----RVVMESDLHDLISRARSSLAVLVS-- 200
+ T LS++LTN TCLDGL+ +S R + L + SLA
Sbjct: 408 SLQGENIQTLLSAILTNQQTCLDGLKDTSSAWSFRNGLTIPLSNDTKLYSVSLAFFTKGW 587
Query: 201 VLPPKANDGFIDEK------LNGDFPSWVTSKDRRLLES----------SVGD---IKAN 241
V P F + K NG P +TSK R + ES VG+ ++
Sbjct: 588 VNPKTNKTSFPNSKHSNKGFKNGRLPLKMTSKTRAIYESVSRRKLLQSNQVGEDVVVRDI 767
Query: 242 VVVAQDGSGKFKTVAQAVASAPDNGKTR---YVIYVKKGTYKENIEIGKKKTNVMLVGDG 298
V V+QDGSG F T+ A+A+AP+ + ++IYV G Y+E I I KKKT +M++GDG
Sbjct: 768 VTVSQDGSGNFTTINDAIAAAPNKSVSSDGYFLIYVTAGVYEEYITIDKKKTYLMMIGDG 947
Query: 299 MDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRVGADQSV 358
++ TIITG+ + +DG TTF S T A VG GF+ ++ +NTAG KHQAVALR GAD S
Sbjct: 948 INKTIITGNHSVVDGWTTFGSPTFAVVGQGFVGVNMTIRNTAGAVKHQAVALRNGADLST 1127
Query: 359 INRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKPMANQKNMV 418
C + +QDTLY HS RQFYR+ I GTVDFIFGNA VVFQ L R PM+ Q N +
Sbjct: 1128FYSCSFEGYQDTLYTHSLRQFYRECDIYGTVDFIFGNAKVVFQNCNLYPRLPMSGQFNAI 1307
Query: 419 TAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTVVLQSVVDGHID 478
TAQGR DPNQ+T TSI C + + DL G++ TYLGRPWK+YSRTV +Q+ +D I+
Sbjct: 1308TAQGRTDPNQDTGTSIHNCTIKATDDLAASNGAVSTYLGRPWKEYSRTVYMQTFMDNVIN 1487
Query: 479 PAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASKFTVTQLIQ 538
AGW WD + L TLYY E+ NSG G+ T RVTW GYH+I NA +A+ FTV +
Sbjct: 1488VAGWRAWD--GEFALSTLYYAEFNNSGPGSSTDGRVTWQGYHVI-NATDAANFTVANFLL 1658
Query: 539 GNVWLKNTGVAFIEGL 554
G+ WL TGV++ L
Sbjct: 1659GDDWLPQTGVSYTNSL 1706
>TC77735 similar to GP|14334646|gb|AAK59501.1 putative pectinesterase
{Arabidopsis thaliana}, partial (59%)
Length = 1963
Score = 339 bits (869), Expect = 2e-93
Identities = 218/560 (38%), Positives = 305/560 (53%), Gaps = 54/560 (9%)
Frame = +2
Query: 49 SSAPNLCEHALDTKSCLTHVSEVVQGSTLSNTKDHKLSTLVSLLTKSTAHIRKAMDTANV 108
SS+ C+ L K C + +S + S+ S+ ++ ++ L + + +D N
Sbjct: 137 SSSSAACKTTLYPKLCRSMLSAI--RSSPSDPYNYGKFSIKQNLKVARKLEKVFIDFLNR 310
Query: 109 IKRRVNSPREEI-ALNDCEELMDLSMDRVWDSVLTLTKNNIDSQHDAHT--------WLS 159
+ + EE+ AL DC++L L++D + +S+ K+ S + T +LS
Sbjct: 311 HQSSSSLNHEEVGALVDCKDLNSLNVDYL-ESISDELKSASSSSSSSDTELVDKIESYLS 487
Query: 160 SVLTNHATCLDGLEGSSRVVMESDLHDLIS------------------------------ 189
+V TNH TC DGL VV +S++ + ++
Sbjct: 488 AVATNHYTCYDGL-----VVTKSNIANALAVPLKDATQFYSVSLGLVTEALSKNMKRNKT 652
Query: 190 ----------RARSSLAVLVSVLPPKANDGFIDEKLNGDFPSWVTSKDRRLLESSVGDIK 239
+ R L L+ +L K + +K + + S T +R L ES I
Sbjct: 653 RKHGLPNKSFKVRQPLEKLIKLLRTK----YSCQKTSSNCTS--TRTERILKESESHGIL 814
Query: 240 AN--VVVAQDGSGKFKTVAQAVASAPDNGKTR---YVIYVKKGTYKENIEIGKKKTNVML 294
N V+V+ G ++ A+A+AP+N K Y+IYV++G Y+E + + K K N++L
Sbjct: 815 LNDFVLVSPYGIANHTSIGDAIAAAPNNTKPEDGYYLIYVREGYYEEYVIVPKHKNNILL 994
Query: 295 VGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRVGA 354
VGDG++ TIITG+ + IDG TTF S+T A G+ FIA DI F+NTAGP+KHQAVA+R A
Sbjct: 995 VGDGINNTIITGNHSVIDGWTTFNSSTFAVSGERFIAVDITFRNTAGPEKHQAVAVRNNA 1174
Query: 355 DQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKPMANQ 414
D S RC + +QDTLY HS RQFYRD I GTVDFIFGNAAVVFQ + ARKP+ NQ
Sbjct: 1175DLSTFYRCSFEGYQDTLYVHSVRQFYRDCKIYGTVDFIFGNAAVVFQNCNIYARKPLPNQ 1354
Query: 415 KNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTVVLQSVVD 474
KN VTAQGR DPNQNT SIQ C + + DL S +YLGRPWK YSRTV +QS +
Sbjct: 1355KNAVTAQGRTDPNQNTGISIQNCTIDAAQDLANDLNSTMSYLGRPWKIYSRTVYMQSYIG 1534
Query: 475 GHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASKFTVT 534
+ P+GW EW+ L T++YGE+ N G G+ T RV WPG H + N +A FTV
Sbjct: 1535DFVQPSGWLEWNGTVG--LDTIFYGEFNNYGPGSVTNNRVQWPG-HFLLNDTQAWNFTVL 1705
Query: 535 QLIQGNVWLKNTGVAFIEGL 554
GN WL +T + + EGL
Sbjct: 1706NFTLGNTWLPDTDIPYTEGL 1765
>NP212791 NP212791|AJ249611.1|CAB65291.1 pectin methyl-esterase PEF1
Length = 1698
Score = 291 bits (745), Expect = 4e-79
Identities = 183/518 (35%), Positives = 270/518 (51%), Gaps = 18/518 (3%)
Frame = +1
Query: 54 LCEHALDTKSCLTHVSEVVQGSTLSNTKDHKLSTLVSLLTKSTAHIRKAMDTANVIKRRV 113
LC+ ++C + ++ ++ SN K+ + L + +RK ++ + + +
Sbjct: 175 LCQSTKFKETC----HKTLEKASFSNMKNR----IKGALGATEEELRKHINNSALYQELA 330
Query: 114 NSPREEIALNDCEELMDLSMDRVWDSVLTLTKNNI----DSQHDAHTWLSSVLTNHATCL 169
+ A+ C E++D ++D + SV TL + + + D WL+ L++ TCL
Sbjct: 331 TDSMTKQAMEICNEVLDYAVDGIHKSVGTLDQFDFHKLSEYAFDIKVWLTGTLSHQQTCL 510
Query: 170 DGLEGSSRVVMESDLHDL-ISRARSSLAVLVSVLPPKANDGF------IDEKLNGD--FP 220
DG + E+ L S SS A+ + + + GF + +L D P
Sbjct: 511 DGFVNTKTHAGETMAKVLKTSMELSSNAIDMMDVVSRILKGFHPSQYGVSRRLLSDDGIP 690
Query: 221 SWVTSKDRRLLESSVGDIKANVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKGTYK 280
SWV+ R LL G++KAN VVAQDGSG+FKT+ A+ + P +VIYVK G YK
Sbjct: 691 SWVSDGHRHLLAG--GNVKANAVVAQDGSGQFKTLTDALKTVPPTNAAPFVIYVKAGVYK 864
Query: 281 ENIEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTA 340
E + + K+ V ++GDG T TGSLN+ DG T+K+AT G F+A+DI F+NTA
Sbjct: 865 ETVNVAKEMNYVTVIGDGPTKTKFTGSLNYADGINTYKTATFGVNGANFMAKDIGFENTA 1044
Query: 341 GPQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVF 400
G K QAVALRV ADQ++ + C++D FQDTL+ S RQFYRD I+GT+DF+FG+A VF
Sbjct: 1045GTSKFQAVALRVTADQAIFHNCQMDGFQDTLFVESQRQFYRDCAISGTIDFVFGDAFGVF 1224
Query: 401 QKSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPW 460
Q KL R P QK +VTA GR+ N +A L V + +YLGRPW
Sbjct: 1225QNCKLICRVPAKGQKCLVTAGGRDKQNSASALVFLSSHFTGEPALTSVTPKL-SYLGRPW 1401
Query: 461 KKYSRTVVLQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYH 520
K YS+ V++ S +D P G+ + F T + EY N G GA T RV W G
Sbjct: 1402KLYSKVVIMDSTIDAMFAPEGYMPMVGGA--FKDTCTFYEYNNKGPGADTNLRVKWHGVK 1575
Query: 521 IIKN--AAE---ASKFTVTQLIQGNVWLKNTGVAFIEG 553
++ + AAE F + + W+ +GV + G
Sbjct: 1576VLTSNVAAEYYPGKFFEIVNATARDTWIVKSGVPYSLG 1689
>TC91752 similar to GP|6630559|gb|AAF19578.1| putative pectinesterase
{Arabidopsis thaliana}, partial (34%)
Length = 691
Score = 260 bits (665), Expect = 8e-70
Identities = 127/213 (59%), Positives = 161/213 (74%), Gaps = 2/213 (0%)
Frame = +3
Query: 220 PSWVTSKDRRLLESSVGDIK--ANVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKG 277
P+W+ KDR+LL+ D+K A++VVA+DGSGK+KT++ A+ P+ R VIYVKKG
Sbjct: 54 PNWLHHKDRKLLQKD-SDLKKKADIVVAKDGSGKYKTISAALKHVPNKSDKRTVIYVKKG 230
Query: 278 TYKENIEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQ 337
Y EN+ + K K NVM++GDGM+ TI++GSLNFIDGT TF +AT A G FIA+DI F+
Sbjct: 231 IYYENVRVEKTKWNVMIIGDGMNVTIVSGSLNFIDGTPTFSTATFAVFGRNFIARDIGFK 410
Query: 338 NTAGPQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAA 397
NTAGPQKHQAVAL ADQ+V +C +DA+QDTLYAHSNRQFYR+ I GTVDFIFGN+A
Sbjct: 411 NTAGPQKHQAVALMTSADQAVFYKCSMDAYQDTLYAHSNRQFYRECNIYGTVDFIFGNSA 590
Query: 398 VVFQKSKLAARKPMANQKNMVTAQGREDPNQNT 430
VV Q + R+PM Q+N +TAQG+ DPN NT
Sbjct: 591 VVLQNCNILPRQPMPGQQNTITAQGKTDPNMNT 689
>BE319906 similar to SP|Q43062|PME_ Pectinesterase PPE8B precursor (EC
3.1.1.11) (Pectin methylesterase) (PE). [Peach] {Prunus
persica}, partial (36%)
Length = 589
Score = 204 bits (519), Expect(2) = 3e-57
Identities = 99/162 (61%), Positives = 126/162 (77%)
Frame = +2
Query: 248 GSGKFKTVAQAVASAPDNGKTRYVIYVKKGTYKENIEIGKKKTNVMLVGDGMDATIITGS 307
GSG + V AV++AP++ RYVIYVKKG Y EN+EI KKK N+ML+G+GMDATII+GS
Sbjct: 2 GSGDYAKVMDAVSAAPESSMKRYVIYVKKGVYVENVEIKKKKWNIMLIGEGMDATIISGS 181
Query: 308 LNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRVGADQSVINRCKIDAF 367
N++DG+TTF+SAT A G GFIA+DI FQNTAG +KHQAVALR +D SV RC I +
Sbjct: 182 RNYVDGSTTFRSATFAVSGRGFIARDISFQNTAGAEKHQAVALRSDSDLSVFYRCGIFGY 361
Query: 368 QDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARK 409
QD+LY H+ RQFYR+ I+GTVDFIFG+A VFQ ++ A++
Sbjct: 362 QDSLYTHTMRQFYRECKISGTVDFIFGDATAVFQNCQILAKE 487
Score = 36.6 bits (83), Expect(2) = 3e-57
Identities = 17/26 (65%), Positives = 19/26 (72%)
Frame = +3
Query: 408 RKPMANQKNMVTAQGREDPNQNTATS 433
+K M QKN VTAQGR+DPNQ T S
Sbjct: 483 KKGMPKQKNTVTAQGRKDPNQPTGFS 560
>BQ138825 similar to PIR|T49922|T49 pectin methylesterase-like protein -
Arabidopsis thaliana, partial (36%)
Length = 658
Score = 203 bits (517), Expect = 1e-52
Identities = 107/218 (49%), Positives = 141/218 (64%), Gaps = 11/218 (5%)
Frame = +1
Query: 257 QAVASAPDNG--KTRYVIYVKKGTYKENIEIGKKKTNVMLVGDGMDATIITGSLNFID-G 313
+AV +APDNG + R+VIY+K+G Y+E + + KK NV+ +GDG+ T+ITGS N G
Sbjct: 4 EAVNAAPDNGVDRKRFVIYIKEGVYEETVRVPLKKRNVVFLGDGIGKTVITGSANVGQPG 183
Query: 314 TTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRVGADQSVINRCKIDAFQDTLYA 373
TT+ SATVA +GDGF+A+D+ +NTAGP HQAVA R+ +D SVI C+ QDTLYA
Sbjct: 184 MTTYNSATVAVLGDGFMAKDLTIENTAGPDAHQAVAFRLDSDLSVIENCEFLGNQDTLYA 363
Query: 374 HSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAAR----KPMANQKNMVTAQGREDPNQN 429
HS RQFY+ I G VDFIFGN+A +FQ ++ R KP + N +TA GR DP Q+
Sbjct: 364 HSLRQFYKSCRIVGNVDFIFGNSAAIFQDCQILVRPRQLKPEKGENNAITAHGRTDPAQS 543
Query: 430 TATSIQQCDVIPSSDLKPVQGS----IKTYLGRPWKKY 463
T Q C + + D + S K YLGRPWK+Y
Sbjct: 544 TGFVFQNCLINGTEDYMALYHSNPKVHKNYLGRPWKEY 657
>BI272853 weakly similar to PIR|T04359|T04 pectin methylesterase-like protein
- maize, partial (25%)
Length = 678
Score = 190 bits (483), Expect = 1e-48
Identities = 97/205 (47%), Positives = 127/205 (61%)
Frame = +1
Query: 269 RYVIYVKKGTYKENIEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDG 328
RY IYVK G Y E I I K N+++ GDG TI+TG N G T ++AT A G
Sbjct: 4 RYTIYVKAGVYDEYITIPKDAVNILMYGDGPGKTIVTGRKNGAAGVKTMQTATFANTALG 183
Query: 329 FIAQDIRFQNTAGPQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGT 388
FI + + F+NTAGP HQAVA R D S + C I +QDTLY +NRQFYR+ I+GT
Sbjct: 184 FIGKAMTFENTAGPAGHQAVAFRNQGDMSALVGCHILGYQDTLYVQTNRQFYRNCVISGT 363
Query: 389 VDFIFGNAAVVFQKSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPV 448
VDFIFG +A + Q S + R P NQ N +TA G NT IQ C+++P + L P
Sbjct: 364 VDFIFGTSATLIQDSTIIVRMPSPNQFNTITADGSYVNKLNTGIVIQGCNIVPKAALFPQ 543
Query: 449 QGSIKTYLGRPWKKYSRTVVLQSVV 473
+ +IK+YLGRPWK ++TVV++S +
Sbjct: 544 RFTIKSYLGRPWKVLTKTVVMESTI 618
>TC84419 similar to SP|Q42920|PME_MEDSA Pectinesterase precursor (EC
3.1.1.11) (Pectin methylesterase) (PE) (P65). [Alfalfa],
partial (53%)
Length = 732
Score = 184 bits (466), Expect = 1e-46
Identities = 96/203 (47%), Positives = 129/203 (63%)
Frame = +2
Query: 329 FIAQDIRFQNTAGPQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGT 388
F A ++ F+N+AG KHQAVALRV AD+++ C+++ +QDTLY S RQFYRD ITGT
Sbjct: 86 FTALNVGFENSAGAAKHQAVALRVTADKALFYNCQMNGYQDTLYTQSKRQFYRDCTITGT 265
Query: 389 VDFIFGNAAVVFQKSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPV 448
+DF+FG+A VFQ KL RKPMANQ+ MVTA GR + +A Q C ++ +
Sbjct: 266 IDFVFGDAVGVFQNCKLIVRKPMANQQCMVTAGGRTKVDSVSALVFQNCHFTGEPEVLTM 445
Query: 449 QGSIKTYLGRPWKKYSRTVVLQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGA 508
Q I YLGRPW+ +S+ V++ S++DG P G+ W F +T Y EY N GAGA
Sbjct: 446 QPKI-AYLGRPWRNFSKVVIVDSLIDGLFVPEGYMPW--MGNLFKETCTYLEYNNKGAGA 616
Query: 509 GTGKRVTWPGYHIIKNAAEASKF 531
T +V WPG I +A EA+K+
Sbjct: 617 ATNLKVKWPGVKTI-SAGEAAKY 682
>TC89457 weakly similar to SP|Q43062|PME_PRUPE Pectinesterase PPE8B
precursor (EC 3.1.1.11) (Pectin methylesterase) (PE).
[Peach] {Prunus persica}, partial (34%)
Length = 1415
Score = 159 bits (401), Expect = 3e-39
Identities = 93/213 (43%), Positives = 135/213 (62%), Gaps = 8/213 (3%)
Frame = +1
Query: 117 REEIALNDCEELMDLSMDRVWDSVLTLTKNNIDSQH------DAHTWLSSVLTNHATCLD 170
R A++DC +L+D+S+D++ S+ K D TWLS+VL TC++
Sbjct: 313 RTSNAVSDCLDLLDMSLDQLNQSISAAQKPKEKDNSTGKLNCDLRTWLSAVLVYPDTCIE 492
Query: 171 GLEGSS-RVVMESDLHDLISRARSSLAVLVSVLPPKANDGFIDEKLNGD-FPSWVTSKDR 228
GLEGS + ++ S L ++S + L +VS ND + N D FPSW+ +D
Sbjct: 493 GLEGSIVKGLISSGLDHVMSLVANLLGEVVS-----GNDDQL--ATNKDRFPSWIRDEDT 651
Query: 229 RLLESSVGDIKANVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKGTYKENIEIGKK 288
+LL+++ + A+ VVA DGSG + V AV++AP++ RYVIYVKKG Y EN+EI KK
Sbjct: 652 KLLQAN--GVTADAVVAADGSGDYAKVMDAVSAAPESSMKRYVIYVKKGVYVENVEIKKK 825
Query: 289 KTNVMLVGDGMDATIITGSLNFIDGTTTFKSAT 321
K N+ML+G+GMDATII+GS N++DG+TTF+SAT
Sbjct: 826 KWNIMLIGEGMDATIISGSRNYVDGSTTFRSAT 924
>TC89627 weakly similar to GP|17529290|gb|AAL38872.1 putative pectin
methylesterase {Arabidopsis thaliana}, partial (33%)
Length = 1289
Score = 119 bits (298), Expect(2) = 3e-38
Identities = 78/234 (33%), Positives = 118/234 (50%), Gaps = 10/234 (4%)
Frame = +3
Query: 239 KANVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKGTYKENIEIGKKKTNVMLVGDG 298
K + V+QDGS +FK++ +A+ S R +I + G Y+E I + K + +GD
Sbjct: 228 KVRLKVSQDGSAQFKSITEALNSIQPYNIRRVIISIAPGYYREKIVVPKTLPFITFLGDV 407
Query: 299 MDATIITG----SLNFIDGTT--TFKSATVAAVGDGFIAQDIRFQNTA----GPQKHQAV 348
D ITG S+ DG TF SATVA F+A +I F+NTA G + QAV
Sbjct: 408 RDPPTITGNDTQSVTGSDGAQLRTFNSATVAVNASYFMAININFENTASFPIGSKVEQAV 587
Query: 349 ALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAAR 408
A+R+ +++ C QDTLY H ++ + I G+VDFI G+ +++ +
Sbjct: 588 AVRITGNKTAFYNCTFSGVQDTLYDHKGLHYFNNCTIKGSVDFICGHGKSLYEGCTI--- 758
Query: 409 KPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKK 462
+ +AN +TAQ +P+ ++ S + VI G TYLGRPW K
Sbjct: 759 RSIANNMTSITAQSGSNPSYDSGFSFKNSMVI---------GDGPTYLGRPWGK 893
Score = 57.8 bits (138), Expect(2) = 3e-38
Identities = 31/81 (38%), Positives = 40/81 (49%)
Frame = +1
Query: 463 YSRTVVLQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHII 522
YS+ V + +D + P GW +W+ + YYGEY SG G+ T RV W +
Sbjct: 895 YSQVVFSYTYMDNSVLPKGWEDWNDTKR--YMNAYYGEYKCSGPGSNTAGRVPWAR---M 1059
Query: 523 KNAAEASKFTVTQLIQGNVWL 543
N EA F TQ I GN WL
Sbjct: 1060LNDKEAQVFIGTQYIDGNTWL 1122
>AL368659 similar to PIR|B86158|B86 hypothetical protein AAF02886.1
[imported] - Arabidopsis thaliana, partial (25%)
Length = 495
Score = 149 bits (375), Expect = 4e-36
Identities = 74/145 (51%), Positives = 99/145 (68%), Gaps = 3/145 (2%)
Frame = +1
Query: 246 QDGSGKFKTVAQAVASAPDNGKTR---YVIYVKKGTYKENIEIGKKKTNVMLVGDGMDAT 302
QDGSG F T+ A+A+AP+N ++I++ +G Y+E + I KKK +M++G+G++ T
Sbjct: 61 QDGSGNFTTINDAIAAAPNNTVASDGYFLIFITEGVYEEYVSIDKKKKYLMMIGEGINQT 240
Query: 303 IITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRVGADQSVINRC 362
IITG+ N DG TTF SAT A V GF+A +I F+NTAG K+QAVALR GAD S C
Sbjct: 241 IITGNRNVADGFTTFNSATFAVVAQGFVAVNITFRNTAGAAKNQAVALRSGADMSTFYSC 420
Query: 363 KIDAFQDTLYAHSNRQFYRDSYITG 387
+ +QDTLY HS RQFYR+ I G
Sbjct: 421 SFEGYQDTLYTHSLRQFYRECDIYG 495
>TC82059 similar to GP|19424045|gb|AAL87311.1 unknown protein {Arabidopsis
thaliana}, partial (17%)
Length = 483
Score = 126 bits (317), Expect = 2e-29
Identities = 58/105 (55%), Positives = 76/105 (72%), Gaps = 1/105 (0%)
Frame = +1
Query: 451 SIKTYLGRPWKKYSRTVVLQSVVDGHIDPAGWAEWDAASKDF-LQTLYYGEYMNSGAGAG 509
++KTYLGRPWK+YSRTV +QS +D I+P+GW EW+ DF L TLYY EY N GAG+
Sbjct: 10 TVKTYLGRPWKEYSRTVYMQSFMDSFINPSGWREWNG---DFALSTLYYAEYDNRGAGSS 180
Query: 510 TGKRVTWPGYHIIKNAAEASKFTVTQLIQGNVWLKNTGVAFIEGL 554
T RVTWPGYH+I A +A+ FTV+ + G+ W+ TGV ++ GL
Sbjct: 181 TANRVTWPGYHVI-GATDAANFTVSNFVSGDDWIPQTGVPYMSGL 312
>AA661011 weakly similar to PIR|T10494|T104 pectinesterase (EC 3.1.1.11)
PECS-c2 - sweet orange, partial (14%)
Length = 649
Score = 122 bits (307), Expect = 3e-28
Identities = 71/167 (42%), Positives = 88/167 (52%), Gaps = 3/167 (1%)
Frame = -3
Query: 391 FIFGNAAVVFQKSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCD--VIPSSDLKPV 448
F GN A + K + + Q NM+ G DP QN S I S K
Sbjct: 647 FXLGNPAX-YSKXETFXXEAXRGQANMIPGPGWGDPFQNXGISFHNVK*GXIRS---KTC 480
Query: 449 QGSIKTYLGRPWKKYSRTVVLQSVVDGHIDPAGWAEWDAASKDFLQ-TLYYGEYMNSGAG 507
G +T LGRPW++YSR +V+++ +D + P GW+ DF Q TLYYGEY N G G
Sbjct: 479 CGQXQTXLGRPWQQYSRVMVMKTFMDTLVSPLGWSP--XGDTDFAQDTLYYGEYENYGPG 306
Query: 508 AGTGKRVTWPGYHIIKNAAEASKFTVTQLIQGNVWLKNTGVAFIEGL 554
+ T RV WPGYH+I N EASKFTV L+ G WL T V F GL
Sbjct: 305 SSTANRVKWPGYHVISNPKEASKFTVAGLLAGPTWLATTTVPFTSGL 165
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.131 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,232,046
Number of Sequences: 36976
Number of extensions: 196281
Number of successful extensions: 1043
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 983
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 990
length of query: 554
length of database: 9,014,727
effective HSP length: 101
effective length of query: 453
effective length of database: 5,280,151
effective search space: 2391908403
effective search space used: 2391908403
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC122166.11


