

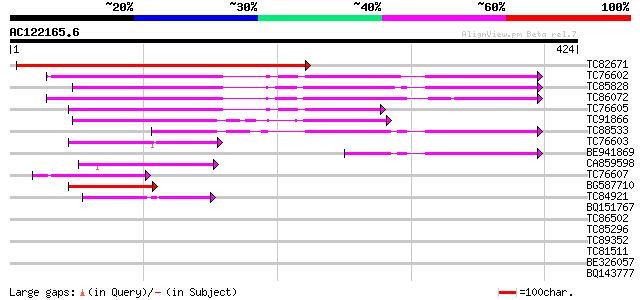
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122165.6 + phase: 0
(424 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC82671 similar to GP|8778327|gb|AAF79336.1| F14J16.13 {Arabidop... 396 e-111
TC76602 similar to SP|Q43317|CYSK_CITLA Cysteine synthase (EC 4.... 158 3e-39
TC85828 homologue to GP|18252506|gb|AAL66291.1 cysteine synthase... 145 2e-35
TC86072 similar to GP|11559260|dbj|BAB18760. beta-cyanoalanine s... 142 3e-34
TC76605 similar to SP|Q43317|CYSK_CITLA Cysteine synthase (EC 4.... 133 1e-31
TC91866 similar to SP|O22682|CYSN_ARATH Probable cysteine syntha... 110 1e-24
TC88533 similar to GP|12081919|dbj|BAB20862. plastidic cysteine ... 99 3e-21
TC76603 similar to GP|18252506|gb|AAL66291.1 cysteine synthase {... 71 7e-13
BE941869 similar to GP|12081919|db plastidic cysteine synthase 1... 66 2e-11
CA859598 similar to GP|15825003|gb| cystathionine beta-synthase ... 59 3e-09
TC76607 similar to GP|18252506|gb|AAL66291.1 cysteine synthase {... 52 4e-07
BG587710 similar to SP|P31300|CYSL Cysteine synthase chloroplas... 50 2e-06
TC84921 similar to PIR|T04211|T04211 probable threonine dehydrat... 42 3e-04
BQ151767 homologue to PIR|S22489|S22 IgE-dependent histamine-rel... 30 2.3
TC86502 similar to PIR|T06786|T06786 6a-hydroxymaackiain methylt... 29 3.0
TC85296 weakly similar to GP|2505870|emb|CAA72908.1 hypothetical... 29 3.9
TC89352 similar to GP|7939581|dbj|BAA95697.1 thionin like protei... 28 6.7
TC81511 similar to GP|20259627|gb|AAM14170.1 unknown protein {Ar... 28 6.7
BE326057 similar to PIR|T46090|T460 hypothetical protein T20E23.... 28 6.7
BQ143777 similar to PIR|T10521|T10 beta-glucosidase (EC 3.2.1.21... 28 6.7
>TC82671 similar to GP|8778327|gb|AAF79336.1| F14J16.13 {Arabidopsis
thaliana}, partial (33%)
Length = 685
Score = 396 bits (1017), Expect = e-111
Identities = 206/221 (93%), Positives = 207/221 (93%), Gaps = 1/221 (0%)
Frame = +1
Query: 6 ARNAGAVGVGTAIFVSLVVAYFFCDRLCNPSKKKKKKKSKNGIIDAIGNTPLIRINSLSD 65
ARNAGAVGVGTAIFVSLVVAYFFCDRLCNPSKKKKKKKSKNGIIDAIGNTPLIRINSLSD
Sbjct: 1 ARNAGAVGVGTAIFVSLVVAYFFCDRLCNPSKKKKKKKSKNGIIDAIGNTPLIRINSLSD 180
Query: 66 ATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRRGGIVTEGSAGSTAISIATVAP 125
ATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRRGGIVTEGSAGSTAISIATVAP
Sbjct: 181 ATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRRGGIVTEGSAGSTAISIATVAP 360
Query: 126 AYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITHKDHFVNIARRRASEANEFAFK 185
AYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITHKDHFVNIARRRASEANEFAFK
Sbjct: 361 AYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITHKDHFVNIARRRASEANEFAFK 540
Query: 186 HRKSQPKGTDSQQINGCKS-DGHKHSTLFPNDCQGGFFADQ 225
HRKSQPKGTDSQQINGCK TL + GFFADQ
Sbjct: 541 HRKSQPKGTDSQQINGCKI*WAQAQHTLS**LSKAGFFADQ 663
>TC76602 similar to SP|Q43317|CYSK_CITLA Cysteine synthase (EC 4.2.99.8)
(O-acetylserine sulfhydrylase) (O-acetylserine
(Thiol)-lyase), partial (97%)
Length = 1355
Score = 158 bits (400), Expect = 3e-39
Identities = 115/373 (30%), Positives = 170/373 (44%), Gaps = 2/373 (0%)
Frame = +2
Query: 28 FCDRLCNPSKKKKKKKSKNGIIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDR 87
FC RL + ++ K + IGNTPL+ +N++++ I K E+L SVKDR
Sbjct: 155 FC-RLSSSDNMEQHFAIKKDVTQLIGNTPLVYLNNITEGCVARIAAKLEYLQSCCSVKDR 331
Query: 88 VAVQIIEEALESGQLRRGG-IVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQI 146
+++ +IE+A G + G ++ E ++G+T I +A++A G K V +P ++E+ I
Sbjct: 332 ISLSMIEDAESKGLITPGKTVLVEPTSGNTGIGLASIAAMRGYKLLVTMPATMSLERKII 511
Query: 147 IEALGATVERVRPVSITHKDHFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDG 206
+ A GA V P KG D G
Sbjct: 512 LRAFGAEVYLTDPA-------------------------------KGID-----GVFQKA 583
Query: 207 HKHSTLFPNDCQGGFFADQFENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAG 266
+ PN + +QFEN AN + HYE TGPEIW+ + G +DA VA GTGGTV G
Sbjct: 584 DELLAKTPNS----YKLNQFENSANPKIHYETTGPEIWKDSGGRVDALVAGIGTGGTVTG 751
Query: 267 VSKFLQEKNPNIKCYLIDPPGSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRV 326
KFL+EKNP+IK Y ++P S + N P + +GIG V
Sbjct: 752 TGKFLKEKNPDIKVYGVEPTESAVLNG-----------------GKPGKHLIQGIGAGIV 880
Query: 327 TKNFAEAKLDGAFRGTDMEAVEMARFLVKNDGLFLGSSSAMNCVGAVRIA-QSIGPGHTI 385
LD + + EA+E A+ L +GL +G SS A+++ + G I
Sbjct: 881 PPILEVDLLDEVIQVSGEEAIETAKLLASKEGLLMGISSGAATAAAIKLGKRPENAGKLI 1060
Query: 386 VTILCDSGMRHLS 398
V I G R+LS
Sbjct: 1061VVIFPSFGERYLS 1099
>TC85828 homologue to GP|18252506|gb|AAL66291.1 cysteine synthase {Glycine
max}, complete
Length = 1202
Score = 145 bits (367), Expect = 2e-35
Identities = 107/353 (30%), Positives = 160/353 (45%), Gaps = 2/353 (0%)
Frame = +3
Query: 48 IIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRRG-G 106
+ + IG TPL+ +N L+D + K E + P SVKDR+ +I +A E G + G
Sbjct: 132 VTELIGKTPLVYLNRLADGCVARVAAKLELMEPCSSVKDRIGYSMIADAEEKGLITPGQS 311
Query: 107 IVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITHKD 166
++ E ++G+T I +A +A A G K + +P ++E+ I+ A GA + P
Sbjct: 312 VLIEPTSGNTGIGLAFMAAAKGYKLIITMPASMSLERRIILLAFGAELVLTDPA------ 473
Query: 167 HFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFADQF 226
KG + G + PN + QF
Sbjct: 474 -------------------------KG-----MKGAVQKAEELLAKTPN----AYILQQF 551
Query: 227 ENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLIDPP 286
EN AN + HYE TGPEIW+ T+G +DAFV+ GTGGT+ G K+L+E+N NIK ++P
Sbjct: 552 ENPANPKVHYETTGPEIWKGTDGKIDAFVSGIGTGGTITGAGKYLKEQNSNIKLIGVEPV 731
Query: 287 GSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAKLDGAFRGTDMEA 346
S V+ G P +GIG V +D + + EA
Sbjct: 732 ES----PVLSG-------------GKPGPHKIQGIGAGFVPGVLEVNLIDEVVQISSDEA 860
Query: 347 VEMARFLVKNDGLFLGSSSAMNCVGAVRIA-QSIGPGHTIVTILCDSGMRHLS 398
+E A+ L +GLF+G SS A++IA + G IV + G R+LS
Sbjct: 861 IETAKLLALKEGLFVGISSGAAAAAAIKIAKRPENAGKLIVVVFPSFGERYLS 1019
>TC86072 similar to GP|11559260|dbj|BAB18760. beta-cyanoalanine synthase
{Solanum tuberosum}, partial (96%)
Length = 1636
Score = 142 bits (357), Expect = 3e-34
Identities = 108/373 (28%), Positives = 164/373 (43%), Gaps = 2/373 (0%)
Frame = +3
Query: 28 FCDRLCNPSKKKKKKKSKNGIIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDR 87
F R+ + K K + IG TPL+ +N +++ G I K E + P S+KDR
Sbjct: 216 FAQRIRDLPKDLPGTNIKKHVSQLIGRTPLVYLNKVTEGCGAYIAVKQEMMQPTASIKDR 395
Query: 88 VAVQIIEEALESGQLRRGG-IVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQI 146
A+ ++E+A + + G I+ E ++G+ IS+A +A G K + +P ++E+
Sbjct: 396 PALAMMEDAEKKNLITPGKTILIEPTSGNMGISLAFMAAMKGYKMVLTMPSYTSLERRVC 575
Query: 147 IEALGATVERVRPVSITHKDHFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDG 206
+ A GA + P KG + G
Sbjct: 576 MRAFGAELILTDPT-------------------------------KG-----MGGTVKKA 647
Query: 207 HKHSTLFPNDCQGGFFADQFENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAG 266
+ PN F QF N AN + H+E TGPEIWE TNG +D FV G+GGTV+G
Sbjct: 648 YDLLESTPN----AFMLQQFSNPANTKVHFETTGPEIWEDTNGQVDIFVMGIGSGGTVSG 815
Query: 267 VSKFLQEKNPNIKCYLIDPPGSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRV 326
V ++L+ +NPN+K Y ++P S + N G P G+G
Sbjct: 816 VGQYLKSQNPNVKIYGVEPSESNVLNGGKPG---------------PHQITGNGVGFKPD 950
Query: 327 TKNFAEAKLDGAFRGTDMEAVEMARFLVKNDGLFLGSSSAMNCVGAVRIAQ-SIGPGHTI 385
+ ++ + +AV MAR L +GL +G SS N V A+R+A G I
Sbjct: 951 ILDM--DVMEKVLEVSSEDAVNMARTLALKEGLMVGISSGANTVAALRLASLPENKGKLI 1124
Query: 386 VTILCDSGMRHLS 398
VT+ G R+LS
Sbjct: 1125VTVHPSFGERYLS 1163
>TC76605 similar to SP|Q43317|CYSK_CITLA Cysteine synthase (EC 4.2.99.8)
(O-acetylserine sulfhydrylase) (O-acetylserine
(Thiol)-lyase), partial (97%)
Length = 1232
Score = 133 bits (334), Expect = 1e-31
Identities = 83/238 (34%), Positives = 118/238 (48%), Gaps = 1/238 (0%)
Frame = +2
Query: 45 KNGIIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRR 104
K + + IGNTPL+ +N++++ I K E+L SVKDR+A+ +IE+A G +
Sbjct: 71 KKDVTELIGNTPLVYLNNITEGCVARIAAKLEYLQSCCSVKDRIALSMIEDAENKGLITP 250
Query: 105 G-GIVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSIT 163
G ++ E ++G+T I +A++A G K V +P ++E+ ++ A GA V P
Sbjct: 251 GKSVLVEPTSGNTGIGLASIAAMRGYKLLVSLPFYVSLERRILLRAFGAEVYLTDPA--- 421
Query: 164 HKDHFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFA 223
KG D G + PN +
Sbjct: 422 ----------------------------KGID-----GVFEKAEELLEKTPNS----YMF 490
Query: 224 DQFENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCY 281
+QFEN AN + HYE TGPEIW T G +DA VA GTGGT+ GV KFL+EKNP IK Y
Sbjct: 491 NQFENPANPKIHYETTGPEIWRDTGGRIDALVAGIGTGGTITGVGKFLKEKNPEIKVY 664
>TC91866 similar to SP|O22682|CYSN_ARATH Probable cysteine synthase
chloroplast precursor (EC 4.2.99.8) (O- acetylserine
sulfhydrylase), partial (64%)
Length = 997
Score = 110 bits (274), Expect = 1e-24
Identities = 74/239 (30%), Positives = 113/239 (46%), Gaps = 1/239 (0%)
Frame = +3
Query: 48 IIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRRGG- 106
+ IGNTP+I +N ++D + K E + P SVKDR+ ++ +A ESG + G
Sbjct: 192 VTQLIGNTPMIYLNKVTDGCVANVAAKLESMEPCRSVKDRIGYSMLADAEESGAISPGKT 371
Query: 107 IVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITHKD 166
++ E + G+T + +A VA G K V +P IE+ ++ GA V +T+ +
Sbjct: 372 VLVEPTTGNTGLGLAFVAATKGYKLIVTMPASVNIERRILLRTFGAEVV------LTNAE 533
Query: 167 HFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFADQF 226
+ A + E + GT PN + QF
Sbjct: 534 KGLKGA---VDKTEEIVY--------GT-------------------PN----AYMFRQF 611
Query: 227 ENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLIDP 285
+N N + H+E TGPEIWE T GN+D VAA GTGGT+ G ++L+ N IK ++P
Sbjct: 612 DNRNNTKIHFETTGPEIWEDTMGNVDLLVAAIGTGGTITGTGQYLKLMNKKIKVVGVEP 788
>TC88533 similar to GP|12081919|dbj|BAB20862. plastidic cysteine synthase 1
{Solanum tuberosum}, partial (65%)
Length = 1136
Score = 99.0 bits (245), Expect = 3e-21
Identities = 76/293 (25%), Positives = 130/293 (43%), Gaps = 1/293 (0%)
Frame = +1
Query: 107 IVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITHKD 166
++ E ++G+T I +A +A + G K + +P ++E+ +++A GA + +T
Sbjct: 22 VLVEPTSGNTGIGLAFIAASKGYKLILTMPASMSLERRVLLKAFGAELV------LTEAA 183
Query: 167 HFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFADQF 226
+N A ++A E KS P + QF
Sbjct: 184 KGMNGAVQKAEEIV-------KSTPDA---------------------------YMLQQF 261
Query: 227 ENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLIDPP 286
+N +N + H+E TGPEIWE T G +D VA GTGGT++G +FL+++N ++ ++P
Sbjct: 262 DNPSNPKIHFETTGPEIWEDTRGKIDILVAGIGTGGTISGTGRFLKQQNSKVQVIGVEP- 438
Query: 287 GSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAKLDGAFRGTDMEA 346
L + ++ G P +GIG V +N E LD + EA
Sbjct: 439 ---LESNILSG-------------GKPGPHKIQGIGAGFVPRNLDEEVLDEVIAISSDEA 570
Query: 347 VEMARFLVKNDGLFLGSSSAMNCVGAVRIA-QSIGPGHTIVTILCDSGMRHLS 398
+E + + +GL +G SS A+++A + G I + G R+LS
Sbjct: 571 IETTKQIALQEGLLVGISSGAAAAAALQVAKRPENEGKLIGVVFPSFGERYLS 729
>TC76603 similar to GP|18252506|gb|AAL66291.1 cysteine synthase {Glycine
max}, partial (42%)
Length = 577
Score = 71.2 bits (173), Expect = 7e-13
Identities = 41/117 (35%), Positives = 67/117 (57%), Gaps = 2/117 (1%)
Frame = +3
Query: 45 KNGIIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRR 104
K + IGNTPL+ +N++++ I K E+L SVKDR+++ +IE+A G +
Sbjct: 165 KKDVTQLIGNTPLVYLNNITEGCVARIAAKLEYLQSCCSVKDRISLSMIEDAESKGLITP 344
Query: 105 G--GIVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRP 159
G G+V E ++G+T I +A++A G K V +P ++E+ I+ A GA V P
Sbjct: 345 GKTGLV-EPTSGNTGIGLASIAAMRGYKLLVTMPATMSLERKIILRAFGAEVYLTDP 512
>BE941869 similar to GP|12081919|db plastidic cysteine synthase 1 {Solanum
tuberosum}, partial (38%)
Length = 607
Score = 66.2 bits (160), Expect = 2e-11
Identities = 45/149 (30%), Positives = 73/149 (48%), Gaps = 1/149 (0%)
Frame = +3
Query: 251 LDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLIDPPGSGLFNKVMRGVMYTKEEAEGRRL 310
+D VA GTGGT++G +FL+++NP IKC ++P L + ++ G
Sbjct: 3 VDILVAGIGTGGTLSGAGRFLKQQNPKIKCIGVEP----LESNILSG------------- 131
Query: 311 KNPFDTITEGIGINRVTKNFAEAKLDGAFRGTDMEAVEMARFLVKNDGLFLGSSSAMNCV 370
P I +GIG V KN + LD + E+V+ A+ + +GL +G SS
Sbjct: 132 GKPRPHIIQGIGAGFVPKNLDKEILDEVIAISGEESVKTAKQIALQEGLLVGISSGCAAT 311
Query: 371 GAVRIA-QSIGPGHTIVTILCDSGMRHLS 398
A+++A + G IV + G R++S
Sbjct: 312 AALQVAKRPENEGKLIVVVFPSFGERYVS 398
>CA859598 similar to GP|15825003|gb| cystathionine beta-synthase {Pichia
pastoris}, partial (4%)
Length = 479
Score = 59.3 bits (142), Expect = 3e-09
Identities = 37/118 (31%), Positives = 66/118 (55%), Gaps = 13/118 (11%)
Frame = +1
Query: 52 IGNTPLIRINSLS------------DATGCEILGKCEFLNP-GGSVKDRVAVQIIEEALE 98
IGNTP++R+N ++ E L K E+ N GS+KDRVA +I+ +A +
Sbjct: 19 IGNTPIVRLNPNKFINTNITEEGHINSKNVEYLVKLEYTNRLSGSIKDRVAKKILSDAEK 198
Query: 99 SGQLRRGGIVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVER 156
S +++ + ++G+ AISIA +A G K ++P+ I++ Q++++LG + R
Sbjct: 199 SCKIKPDTTLLIPTSGNLAISIALLAQRRGYKTIALVPERTTIDRIQLLKSLGIEIIR 372
>TC76607 similar to GP|18252506|gb|AAL66291.1 cysteine synthase {Glycine
max}, partial (53%)
Length = 608
Score = 52.0 bits (123), Expect = 4e-07
Identities = 29/88 (32%), Positives = 48/88 (53%)
Frame = +3
Query: 18 IFVSLVVAYFFCDRLCNPSKKKKKKKSKNGIIDAIGNTPLIRINSLSDATGCEILGKCEF 77
+F SL+ FC RL + ++ K + IGNTPL+ +N++++ I K E+
Sbjct: 15 VFNSLLCFERFC-RLSSSDNMEQHFAIKKDVTQLIGNTPLVYLNNITEGCVARIAAKLEY 191
Query: 78 LNPGGSVKDRVAVQIIEEALESGQLRRG 105
L SVKDR+++ +IE+A G + G
Sbjct: 192 LQSCCSVKDRISLSMIEDAESKGLITPG 275
Score = 33.9 bits (76), Expect = 0.12
Identities = 13/22 (59%), Positives = 16/22 (72%)
Frame = +3
Query: 241 PEIWEQTNGNLDAFVAAAGTGG 262
PEIW+ + G +DA VA GTGG
Sbjct: 543 PEIWKDSGGRVDAMVAGIGTGG 608
Score = 27.7 bits (60), Expect = 8.7
Identities = 12/27 (44%), Positives = 17/27 (62%), Gaps = 4/27 (14%)
Frame = +2
Query: 225 QFENLANFRAHYEGTGP----EIWEQT 247
+FEN AN + HYE TGP +W ++
Sbjct: 494 RFENSANPKIHYETTGP*DLERLWRES 574
>BG587710 similar to SP|P31300|CYSL Cysteine synthase chloroplast precursor
(EC 4.2.99.8) (O-acetylserine sulfhydrylase), partial
(31%)
Length = 804
Score = 50.1 bits (118), Expect = 2e-06
Identities = 22/66 (33%), Positives = 40/66 (60%)
Frame = +1
Query: 45 KNGIIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRR 104
K + IGNTPL+ +N++++ I K E+L SVKDR+++ +IE+A G +
Sbjct: 10 KKDVTQLIGNTPLVYLNNITEGCVARIAAKLEYLQSCCSVKDRISLSMIEDAESKGLITP 189
Query: 105 GGIVTE 110
G ++++
Sbjct: 190 GKVISQ 207
>TC84921 similar to PIR|T04211|T04211 probable threonine dehydratase (EC
4.2.1.16) T5C23.70 [similarity] - Arabidopsis thaliana,
partial (32%)
Length = 696
Score = 42.4 bits (98), Expect = 3e-04
Identities = 28/100 (28%), Positives = 50/100 (50%)
Frame = +2
Query: 55 TPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRRGGIVTEGSAG 114
TP++ SL+ +G ++ KCE GG+ K R A + + + G++T S+G
Sbjct: 143 TPVLSSTSLNAISGRQLYFKCESFQKGGAFKFRGACNAVFSLNDEDASK--GVITH-SSG 313
Query: 115 STAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATV 154
+ A ++A A G ++VIP +A K + ++ G V
Sbjct: 314 NHAAALALAAKLRGIPAYIVIPKNAPTCKIENVKRYGGQV 433
>BQ151767 homologue to PIR|S22489|S22 IgE-dependent histamine-releasing
factor homolog - alfalfa (fragment), partial (76%)
Length = 719
Score = 29.6 bits (65), Expect = 2.3
Identities = 16/32 (50%), Positives = 19/32 (59%)
Frame = +2
Query: 14 VGTAIFVSLVVAYFFCDRLCNPSKKKKKKKSK 45
V T I+V L++ FC L P K KKKKK K
Sbjct: 485 VXTMIWVFLIMMX*FCPYLSFPIKFKKKKKKK 580
>TC86502 similar to PIR|T06786|T06786 6a-hydroxymaackiain methyltransferase
(EC 2.1.1.-) - garden pea, partial (98%)
Length = 1256
Score = 29.3 bits (64), Expect = 3.0
Identities = 15/36 (41%), Positives = 22/36 (60%)
Frame = +1
Query: 251 LDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLIDPP 286
L++ V AG G V SK + E+ P+IKC ++D P
Sbjct: 652 LESLVDVAGGTGVV---SKLIHEEFPHIKCTVLDQP 750
>TC85296 weakly similar to GP|2505870|emb|CAA72908.1 hypothetical protein
{Arabidopsis thaliana}, partial (3%)
Length = 774
Score = 28.9 bits (63), Expect = 3.9
Identities = 13/24 (54%), Positives = 15/24 (62%)
Frame = +1
Query: 26 YFFCDRLCNPSKKKKKKKSKNGII 49
Y+ C LCN KKKKKKK + I
Sbjct: 439 YYPCPILCNTYKKKKKKKFLSSFI 510
>TC89352 similar to GP|7939581|dbj|BAA95697.1 thionin like protein
{Nicotiana tabacum}, partial (64%)
Length = 499
Score = 28.1 bits (61), Expect = 6.7
Identities = 17/51 (33%), Positives = 29/51 (56%), Gaps = 3/51 (5%)
Frame = +3
Query: 17 AIFVSLVVAYFFC--DRLCNPSKKKKK-KKSKNGIIDAIGNTPLIRINSLS 64
A +++ V+ + C +CN S KKK KKS +ID + + ++ IN +S
Sbjct: 345 ATYLASVIIIYLCIFSFMCNQSSLKKK*KKSVTNLIDIVIS*YIVTINLVS 497
>TC81511 similar to GP|20259627|gb|AAM14170.1 unknown protein {Arabidopsis
thaliana}, partial (41%)
Length = 725
Score = 28.1 bits (61), Expect = 6.7
Identities = 11/32 (34%), Positives = 19/32 (59%)
Frame = -2
Query: 27 FFCDRLCNPSKKKKKKKSKNGIIDAIGNTPLI 58
++ + +C+P+K K KK KNG I P++
Sbjct: 655 YYTNVMCSPTKNKSKKTQKNGKIIGTQFLPIL 560
>BE326057 similar to PIR|T46090|T460 hypothetical protein T20E23.190 -
Arabidopsis thaliana, partial (13%)
Length = 629
Score = 28.1 bits (61), Expect = 6.7
Identities = 17/52 (32%), Positives = 26/52 (49%), Gaps = 2/52 (3%)
Frame = +2
Query: 346 AVEMARFLVKNDGLFLGSSSAMNCVGAVRIAQSIGP--GHTIVTILCDSGMR 395
AV + ++V+ D L S +GA + S P GHT++ IL D +R
Sbjct: 347 AVAIGTYIVEFDALTWKQDSPPLDIGAPAVRMSYSPTSGHTVIAILQDCTIR 502
>BQ143777 similar to PIR|T10521|T10 beta-glucosidase (EC 3.2.1.21) - common
nasturtium, partial (17%)
Length = 750
Score = 28.1 bits (61), Expect = 6.7
Identities = 15/40 (37%), Positives = 24/40 (59%), Gaps = 4/40 (10%)
Frame = +2
Query: 22 LVVAYFFCDRLC----NPSKKKKKKKSKNGIIDAIGNTPL 57
++V +++CD LC +PSKKK+KK + N+PL
Sbjct: 440 MLVPHWYCDILCLKYFSPSKKKQKKTGGGA---PVPNSPL 550
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,636,745
Number of Sequences: 36976
Number of extensions: 150931
Number of successful extensions: 1144
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 1030
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1116
length of query: 424
length of database: 9,014,727
effective HSP length: 99
effective length of query: 325
effective length of database: 5,354,103
effective search space: 1740083475
effective search space used: 1740083475
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC122165.6


