

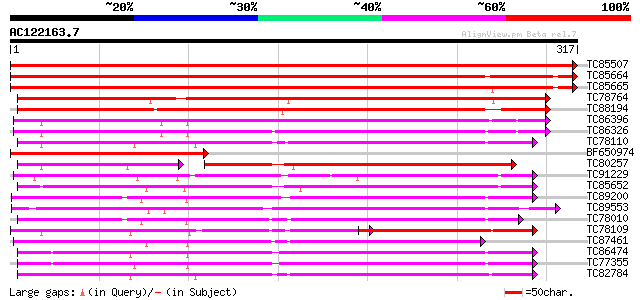
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122163.7 - phase: 0
(317 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85507 homologue to SP|P31239|ACCO_PEA 1-aminocyclopropane-1-ca... 644 0.0
TC85664 1-aminocyclopropanecarboxylic acid oxidase [Medicago tru... 514 e-146
TC85665 similar to GP|18157333|dbj|BAB83762. 1-aminocyclopropane... 491 e-139
TC78764 similar to PIR|C96802|C96802 hypothetical protein F2P24.... 304 3e-83
TC88194 similar to PIR|F84578|F84578 1-aminocyclopropane-1-carbo... 291 3e-79
TC86396 similar to GP|10178137|dbj|BAB11549. leucoanthocyanidin ... 209 1e-54
TC86326 similar to GP|6016680|gb|AAF01507.1| putative leucoantho... 206 1e-53
TC78110 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 193 8e-50
BF650974 similar to SP|P31239|ACCO 1-aminocyclopropane-1-carboxy... 187 6e-48
TC80257 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 141 2e-46
TC91229 similar to GP|9294689|dbj|BAB03055.1 gene_id:MHC9.10~sim... 174 5e-44
TC85652 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 173 9e-44
TC89200 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 171 3e-43
TC89553 weakly similar to SP|P24397|HY6H_HYONI Hyoscyamine 6-dio... 170 6e-43
TC78010 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 169 1e-42
TC78109 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 118 1e-42
TC87461 weakly similar to GP|2688828|gb|AAB88878.1| ethylene-for... 169 1e-42
TC86474 weakly similar to GP|6016680|gb|AAF01507.1| putative leu... 165 2e-41
TC77355 weakly similar to PIR|T05552|T05552 SRG1 protein-related... 162 2e-40
TC82784 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 162 2e-40
>TC85507 homologue to SP|P31239|ACCO_PEA 1-aminocyclopropane-1-carboxylate
oxidase (EC 1.-.-.-) (ACC oxidase) (Ethylene-forming
enzyme) (EFE), complete
Length = 1342
Score = 644 bits (1662), Expect = 0.0
Identities = 317/317 (100%), Positives = 317/317 (100%)
Frame = +1
Query: 1 MENFPVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKC 60
MENFPVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKC
Sbjct: 97 MENFPVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKC 276
Query: 61 MEQRFKEMVASKGLECVQSEINDLDWESTFFLRHLPSSNISEIPDLDEDYRKTMKEFAEK 120
MEQRFKEMVASKGLECVQSEINDLDWESTFFLRHLPSSNISEIPDLDEDYRKTMKEFAEK
Sbjct: 277 MEQRFKEMVASKGLECVQSEINDLDWESTFFLRHLPSSNISEIPDLDEDYRKTMKEFAEK 456
Query: 121 LEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKGLRAHTDA 180
LEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKGLRAHTDA
Sbjct: 457 LEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKGLRAHTDA 636
Query: 181 GGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVMHRVIAQT 240
GGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVMHRVIAQT
Sbjct: 637 GGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVMHRVIAQT 816
Query: 241 DGARMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDYMKLYMGLKFQAKEPRFE 300
DGARMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDYMKLYMGLKFQAKEPRFE
Sbjct: 817 DGARMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDYMKLYMGLKFQAKEPRFE 996
Query: 301 AMMKAMSSVEVGPVVTI 317
AMMKAMSSVEVGPVVTI
Sbjct: 997 AMMKAMSSVEVGPVVTI 1047
>TC85664 1-aminocyclopropanecarboxylic acid oxidase [Medicago truncatula]
Length = 1832
Score = 514 bits (1325), Expect = e-146
Identities = 247/317 (77%), Positives = 279/317 (87%)
Frame = +1
Query: 1 MENFPVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKC 60
M+NFP++++ LN +ERKATME IKDACENWGFFE VNH I +LMD VE+LTKEHY+KC
Sbjct: 637 MDNFPIINLENLNGDERKATMEKIKDACENWGFFELVNHGIPHDLMDTVERLTKEHYRKC 816
Query: 61 MEQRFKEMVASKGLECVQSEINDLDWESTFFLRHLPSSNISEIPDLDEDYRKTMKEFAEK 120
MEQRFKE+V+SKGLE VQ+E+ D+DWESTF LRHLP SNISEIPDL ++YRK+MKEFA K
Sbjct: 817 MEQRFKELVSSKGLEAVQTEVKDMDWESTFHLRHLPESNISEIPDLSDEYRKSMKEFALK 996
Query: 121 LEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKGLRAHTDA 180
LE LAEELLDLLCENLGLEKGYLKK YGSKGP FGTKV+NYPPCPKPDL+KGLRAHTDA
Sbjct: 997 LETLAEELLDLLCENLGLEKGYLKKALYGSKGPTFGTKVANYPPCPKPDLVKGLRAHTDA 1176
Query: 181 GGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVMHRVIAQT 240
GGIILLFQDDKVSGLQLLKD W+DVPPM HSIVINLGDQLEVITNGKYKSV HRV+AQT
Sbjct: 1177GGIILLFQDDKVSGLQLLKDGNWVDVPPMHHSIVINLGDQLEVITNGKYKSVEHRVVAQT 1356
Query: 241 DGARMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDYMKLYMGLKFQAKEPRFE 300
DG RMSIASFYNPG+DAVI PA TL+ +E +EIYPKF+F+DYM LY LKFQAKEPRFE
Sbjct: 1357DGTRMSIASFYNPGSDAVIYPAPTLI--EENNEIYPKFVFEDYMNLYARLKFQAKEPRFE 1530
Query: 301 AMMKAMSSVEVGPVVTI 317
A + S+V +GP+ T+
Sbjct: 1531AFKE--SNVNLGPIATV 1575
>TC85665 similar to GP|18157333|dbj|BAB83762.
1-aminocyclopropane-1-carboxylic acid oxidase {Phaseolus
lunatus}, partial (97%)
Length = 1284
Score = 491 bits (1264), Expect = e-139
Identities = 233/319 (73%), Positives = 274/319 (85%), Gaps = 2/319 (0%)
Frame = +2
Query: 1 MENFPVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKC 60
M NFP++ + KLN ERK TME IKDACENWGFFE VNH I +LMD +E+LTKEHY+KC
Sbjct: 47 MMNFPIISLEKLNGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKC 226
Query: 61 MEQRFKEMVASKGLECVQSEINDLDWESTFFLRHLPSSNISEIPDLDEDYRKTMKEFAEK 120
ME RFKE+++SKGL+ VQ+E+ D+DWESTF +RHLP SNISEIPDL ++YRK MKEF+ +
Sbjct: 227 MEHRFKEVISSKGLDVVQTEVKDMDWESTFHVRHLPESNISEIPDLSDEYRKVMKEFSLR 406
Query: 121 LEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKGLRAHTDA 180
LEKLAEELLDLLCENLGLEKGYLKK FYGS+GP FGTKV+NYP CP P+L+KGLRAHTDA
Sbjct: 407 LEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKGLRAHTDA 586
Query: 181 GGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVMHRVIAQT 240
GGIILLFQDDKVSGLQLLKD QW+DVPPM HSIV+NLGDQLEVITNGKYKSV HRVIAQT
Sbjct: 587 GGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEHRVIAQT 766
Query: 241 DGARMSIASFYNPGNDAVISPASTLLKE--DETSEIYPKFIFDDYMKLYMGLKFQAKEPR 298
+G RMSIASFYNPG+DAVI PA LL++ +E +YPKF+F++YMK+Y LKF AKEPR
Sbjct: 767 NGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMKIYAALKFHAKEPR 946
Query: 299 FEAMMKAMSSVEVGPVVTI 317
FEA+ + S+V +GP+ +
Sbjct: 947 FEALKE--SNVNLGPIAIV 997
>TC78764 similar to PIR|C96802|C96802 hypothetical protein F2P24.4
[imported] - Arabidopsis thaliana, partial (88%)
Length = 1276
Score = 304 bits (779), Expect = 3e-83
Identities = 153/307 (49%), Positives = 207/307 (66%), Gaps = 9/307 (2%)
Frame = +1
Query: 5 PVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKCMEQR 64
PV+D KLN EER T+ I + CE WGFF+ +NH IS EL+++V+K++ E YK E+
Sbjct: 76 PVIDFSKLNGEERAKTLAQIANGCEEWGFFQLINHGISEELLERVKKVSSEFYKLEREEN 255
Query: 65 FKEMVASKGLECV-----QSEINDLDWESTFFLRHLPSSNISEIPDLDEDYRKTMKEFAE 119
FK K L + ++ ++DWE L + +E P+ +R+TM E+
Sbjct: 256 FKNSKTVKLLNDIAEKKSSEKLENVDWEDVITLL-----DDNEWPENTPSFRETMSEYRS 420
Query: 120 KLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPN--FGTKVSNYPPCPKPDLIKGLRAH 177
+L+KLA L +++ ENLGL KGY+KK +G N FGTKVS+YPPCP P+L+ GLRAH
Sbjct: 421 ELKKLAVSLTEVMDENLGLPKGYIKKALNDGEGDNAFFGTKVSHYPPCPHPELVNGLRAH 600
Query: 178 TDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVMHRVI 237
TDAGG+ILLFQDDKV GLQ+LKD +W+DV P+P++IVIN GDQ+EV++NG+ KS HRV+
Sbjct: 601 TDAGGVILLFQDDKVGGLQMLKDGEWLDVQPLPNAIVINTGDQIEVLSNGRNKSCWHRVL 780
Query: 238 AQTDGARMSIASFYNPGNDAVISPASTLLKED--ETSEIYPKFIFDDYMKLYMGLKFQAK 295
T+G R +IASFYNP A ISPA L ++D + + YPKF+F DYM +Y KF K
Sbjct: 781 NFTEGTRRTIASFYNPPLKATISPAPQLAEKDNQQVDDTYPKFVFGDYMSVYAEQKFLPK 960
Query: 296 EPRFEAM 302
EPRF A+
Sbjct: 961 EPRFRAV 981
>TC88194 similar to PIR|F84578|F84578 1-aminocyclopropane-1-carboxylate
oxidase [imported] - Arabidopsis thaliana, partial (91%)
Length = 1018
Score = 291 bits (744), Expect = 3e-79
Identities = 146/302 (48%), Positives = 209/302 (68%), Gaps = 4/302 (1%)
Frame = +2
Query: 5 PVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKCMEQR 64
PV+D LN ++R TM ++ +AC+ WG F NH I +LM+KV+K+ +Y++ +++
Sbjct: 11 PVIDFSTLNGDKRGETMALLHEACQKWGCFLIENHDIEGKLMEKVKKVINSYYEENLKES 190
Query: 65 FKEMVASKGLECVQSEINDLDWESTFFLRHLPSSNISEIPDLDEDYRKTMKEFAEKLEKL 124
F + +K LE ++ D+DWES+FF+ H P+SNI +IP+L ED +TM E+ +KL ++
Sbjct: 191 FYQSEIAKRLEKKENTC-DVDWESSFFIWHRPTSNIRKIPNLSEDLCQTMDEYIDKLVQV 367
Query: 125 AEELLDLLCENLGLEKGYLKKVFYGSK--GPNFGTKVSNYPPCPKPDLIKGLRAHTDAGG 182
AE L ++ ENLGLEK Y+KK F G+ GP GTKV+ YP CP P+L++GLR HTDAGG
Sbjct: 368 AETLSQMMSENLGLEKDYIKKAFSGNNNNGPAMGTKVAKYPECPYPELVRGLREHTDAGG 547
Query: 183 IILLFQDDKVSGLQLLKDDQWIDVPPMP-HSIVINLGDQLEVITNGKYKSVMHRVIAQTD 241
IILL QDDKV GL+ KD +WI++PP ++I +N GDQ+EV++NG YKS +HRV+ +
Sbjct: 548 IILLLQDDKVPGLEFFKDGKWIEIPPSKNNAIFVNTGDQIEVLSNGLYKSAVHRVMPDKN 727
Query: 242 GARMSIASFYNPGNDAVISPASTLLKEDETSEIYP-KFIFDDYMKLYMGLKFQAKEPRFE 300
G+R+SIASFYNP +A+ISPA LL YP + + DY++LY KF K PRFE
Sbjct: 728 GSRLSIASFYNPVGEAIISPAPKLL--------YPSNYCYGDYLELYGKTKFGDKGPRFE 883
Query: 301 AM 302
++
Sbjct: 884 SI 889
>TC86396 similar to GP|10178137|dbj|BAB11549. leucoanthocyanidin
dioxygenase-like protein {Arabidopsis thaliana}, partial
(82%)
Length = 1326
Score = 209 bits (532), Expect = 1e-54
Identities = 113/310 (36%), Positives = 182/310 (58%), Gaps = 10/310 (3%)
Frame = +2
Query: 3 NFPVVDMGKLNTEE---RKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKK 59
N PV+D+ L++E+ R+ ++ + +AC WGFF+ VNH IS ELM+ +++ +E +
Sbjct: 251 NIPVIDLEHLSSEDPVLRETVLKRVSEACREWGFFQVVNHGISHELMESAKEVWREFFNL 430
Query: 60 CMEQRFKEMVASKGLECVQSEIND-----LDWESTFFLRHLPSS--NISEIPDLDEDYRK 112
+E + + + E S + LDW FFL +P S N ++ P RK
Sbjct: 431 PLEVKEEFANSPSTYEGYGSRLGVKKGAILDWSDYFFLHSMPPSLRNQAKWPATPSSLRK 610
Query: 113 TMKEFAEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIK 172
+ E+ E++ KL +L+L+ NLGL++ YL F G +V+ YP CP+PDL
Sbjct: 611 IIAEYGEEVVKLGGRMLELMSTNLGLKEDYLMNAFGGENELGACLRVNFYPKCPQPDLTL 790
Query: 173 GLRAHTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSV 232
GL H+D GG+ +L DD VSGLQ+ K + WI V P+P++ +IN+GDQ++V++N YKSV
Sbjct: 791 GLSPHSDPGGMTILLPDDFVSGLQVRKGNDWITVKPVPNAFIINIGDQIQVLSNAIYKSV 970
Query: 233 MHRVIAQTDGARMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDYMKLYMGLKF 292
HRVI + R+S+A FYNP +D +I PA L+ + E +Y +D+Y +L++ +K
Sbjct: 971 EHRVIVNSIKDRVSLAMFYNPKSDLLIEPAKELVTK-ERPALYSAMTYDEY-RLFIRMKG 1144
Query: 293 QAKEPRFEAM 302
+ + E++
Sbjct: 1145PCGKTQVESL 1174
>TC86326 similar to GP|6016680|gb|AAF01507.1| putative leucoanthocyanidin
dioxygenase {Arabidopsis thaliana}, partial (81%)
Length = 1686
Score = 206 bits (524), Expect = 1e-53
Identities = 112/310 (36%), Positives = 180/310 (57%), Gaps = 10/310 (3%)
Frame = +2
Query: 3 NFPVVDMGKLNTEE---RKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKK 59
N P++D+G LN ++ + ++ I DAC +WGFF+ VNH +S +LMDK + ++ +
Sbjct: 395 NIPIIDLGGLNGDDLDVHASILKQISDACRDWGFFQIVNHGVSPDLMDKARETWRQFFHL 574
Query: 60 CMEQRFKEMVASKGLECVQSEIND-----LDWESTFFLRHLPSS--NISEIPDLDEDYRK 112
ME + + + E S + LDW +FL +LP S + ++ P + R+
Sbjct: 575 PMEAKQQYANSPTTYEGYGSRLGVEKGAILDWSDYYFLHYLPVSVKDCNKWPASPQSCRE 754
Query: 113 TMKEFAEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIK 172
E+ ++L KL+ L+ L NLGLE+ L+ F G + +V+ YP CP+P+L
Sbjct: 755 VFDEYGKELVKLSGRLMKALSLNLGLEEKILQNAF-GGEEIGACMRVNFYPKCPRPELTL 931
Query: 173 GLRAHTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSV 232
GL +H+D GG+ +L DD+V+GLQ+ K D WI V P H ++N+GDQ++V++N YKSV
Sbjct: 932 GLSSHSDPGGMTMLLPDDQVAGLQVRKFDNWITVNPARHGFIVNIGDQIQVLSNATYKSV 1111
Query: 233 MHRVIAQTDGARMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDYMKLYMGLKF 292
HRVI +D R+S+A FYNP +D I P L+ E +YP FD+Y +L++ ++
Sbjct: 1112EHRVIVNSDQERLSLAFFYNPRSDIPIEPLKQLI-TPERPALYPAMTFDEY-RLFIRMRG 1285
Query: 293 QAKEPRFEAM 302
+ + E+M
Sbjct: 1286PCGKSQVESM 1315
>TC78110 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (53%)
Length = 1303
Score = 193 bits (490), Expect = 8e-50
Identities = 102/301 (33%), Positives = 174/301 (56%), Gaps = 10/301 (3%)
Frame = +2
Query: 5 PVVDMGKLNTEE---RKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKCM 61
PV+D KL +++ + ++ + AC+ WGFF+ +NH +S L++ V+ KE Y +
Sbjct: 212 PVIDFSKLFSQDLTIKGLELDKLHSACKEWGFFQLINHGVSTSLVENVKMGAKEFYNLPI 391
Query: 62 EQRFKEM-----VASKGLECVQSEINDLDWESTFFLRHLPSSNISE--IPDLDEDYRKTM 114
E++ K V G V SE LDW FF+ LPS P L +R +
Sbjct: 392 EEKKKFSQKEGDVEGYGQAFVMSEEQKLDWADMFFMITLPSHMRKPHLFPKLPLPFRDDL 571
Query: 115 KEFAEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKGL 174
+ ++ +L+KLA +++D + L ++ ++++F +G T+++ YPPCP+P+L+ GL
Sbjct: 572 ETYSAELKKLAIQIIDFMANALKVDAKEIRELF--GEGTQ-STRINYYPPCPQPELVIGL 742
Query: 175 RAHTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVMH 234
+H+D GG+ +L Q +++ GLQ+ KD WI V P+P++ +INLGD LE+ITNG Y S+ H
Sbjct: 743 NSHSDGGGLTILLQGNEMDGLQIKKDGFWIPVKPLPNAFIINLGDMLEIITNGIYPSIEH 922
Query: 235 RVIAQTDGARMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDYMKLYMGLKFQA 294
R R+SIA+FY+P + ++ P+ TL+ +T ++ D+ K Y+G + +
Sbjct: 923 RATVNLKKERLSIATFYSPSSAVILRPSPTLV-TPKTPALFKPIGVTDFYKGYLGKELRG 1099
Query: 295 K 295
K
Sbjct: 1100K 1102
>BF650974 similar to SP|P31239|ACCO 1-aminocyclopropane-1-carboxylate oxidase
(EC 1.-.-.-) (ACC oxidase) (Ethylene-forming enzyme)
(EFE), partial (40%)
Length = 582
Score = 187 bits (474), Expect = 6e-48
Identities = 91/111 (81%), Positives = 96/111 (85%)
Frame = +1
Query: 1 MENFPVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKC 60
ME FPVVDM LNTEE+ +TME+IKDACENWGFFE VNH ISIE MDKVEKLTKE+YKK
Sbjct: 193 MEKFPVVDMKNLNTEEKASTMEIIKDACENWGFFELVNHGISIEFMDKVEKLTKENYKKR 372
Query: 61 MEQRFKEMVASKGLECVQSEINDLDWESTFFLRHLPSSNISEIPDLDEDYR 111
MEQR KEMVASKGL QSEINDLDWESTFFLRHLP+ NIS IPDLD DYR
Sbjct: 373 MEQRXKEMVASKGLXFAQSEINDLDWESTFFLRHLPNFNISXIPDLDHDYR 525
Score = 39.7 bits (91), Expect = 0.002
Identities = 18/19 (94%), Positives = 18/19 (94%)
Frame = +2
Query: 169 DLIKGLRAHTDAGGIILLF 187
DLIKGLR HTDAGGIILLF
Sbjct: 524 DLIKGLRXHTDAGGIILLF 580
>TC80257 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (50%)
Length = 1376
Score = 141 bits (355), Expect(2) = 2e-46
Identities = 63/177 (35%), Positives = 115/177 (64%), Gaps = 3/177 (1%)
Frame = +2
Query: 110 YRKTMKEFAEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGT---KVSNYPPCP 166
+R+ ++ ++ +LEKLA ++++L+ L + + ++F N GT +V+ YPPCP
Sbjct: 605 FRENLEVYSVELEKLAIKVIELMANALAINPKEMTELF------NIGTQMVRVNYYPPCP 766
Query: 167 KPDLIKGLRAHTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITN 226
+P+ + GL++H+DAGG+ +L Q + GLQ+ KD QWI V P+P++ ++N+GD LE+ITN
Sbjct: 767 QPERVIGLKSHSDAGGLTILLQTSDIDGLQIRKDGQWIPVKPLPNAFIVNIGDMLEIITN 946
Query: 227 GKYKSVMHRVIAQTDGARMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDY 283
G Y S+ HR ++ R+S+A+FY P A+++PA +L+ ++ ++ + D +
Sbjct: 947 GIYPSIEHRATVNSEKERISVAAFYGPNMQAMLAPAPSLVTQERPAQFRRISVVDHF 1117
Score = 62.4 bits (150), Expect(2) = 2e-46
Identities = 34/101 (33%), Positives = 57/101 (55%), Gaps = 8/101 (7%)
Frame = +1
Query: 5 PVVDMGKLNTEE---RKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKCM 61
PV+D+ +L +++ ++ +E + AC+ WGFF+ VNH IS L++ ++K K ++ M
Sbjct: 259 PVIDLAELLSQDINLKRHELEKLHYACKEWGFFQLVNHGISTSLVEDMKKGAKTLFELSM 438
Query: 62 EQR-----FKEMVASKGLECVQSEINDLDWESTFFLRHLPS 97
E++ + + G V SE L+W TFFL LPS
Sbjct: 439 EEKKNLWQIEGEMEGFGQSFVLSEEQKLEWADTFFLSTLPS 561
>TC91229 similar to GP|9294689|dbj|BAB03055.1 gene_id:MHC9.10~similar to
oxidases {Arabidopsis thaliana}, partial (86%)
Length = 1198
Score = 174 bits (440), Expect = 5e-44
Identities = 104/307 (33%), Positives = 169/307 (54%), Gaps = 14/307 (4%)
Frame = +2
Query: 3 NFPVVDMGKL---NTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKK 59
+ PV+D K N EE + + ACE WGFF+ +NH I + LM+ +E +++E +
Sbjct: 161 DMPVIDFSKFSKGNKEEVFNELSKLSTACEEWGFFQVINHEIDLNLMENIEDMSREFFML 340
Query: 60 CMEQRFKEMVA-----SKGLECVQSEINDLDWESTFFL----RHLPSSNISEIPDLDEDY 110
++++ K +A G V SE LDW + F L ++ +SN+ P
Sbjct: 341 PLDEKQKYPMAPGTVQGYGQAFVFSEDQKLDWCNMFALGIEPHYIRNSNLW--PKKPARL 514
Query: 111 RKTMKEFAEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDL 170
+T++ ++ K+ KL + LL + L LE+ +K+F + +++ YP C +PDL
Sbjct: 515 SETIELYSRKIRKLCQNLLKYIALGLSLEEDVFEKMFGEAVQ---AIRMNYYPTCSRPDL 685
Query: 171 IKGLRAHTDAGGIILLFQDDKVS--GLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGK 228
+ GL H+D G + + Q K S GLQ+LKD+ W+ V P+ +++VIN+GD +EV+TNGK
Sbjct: 686 VLGLSPHSD-GSALTVLQQAKGSPVGLQILKDNTWVPVEPISNALVINIGDTIEVLTNGK 862
Query: 229 YKSVMHRVIAQTDGARMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDYMKLYM 288
YKSV HR +A + R+SI +FY P D + P L+ E+ + Y ++ +Y K Y+
Sbjct: 863 YKSVEHRAVAHEEKDRLSIVTFYAPSYDLELGPMLELVDENHPCK-YKRYNHGEYSKHYV 1039
Query: 289 GLKFQAK 295
K Q K
Sbjct: 1040TNKLQGK 1060
>TC85652 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (50%)
Length = 1489
Score = 173 bits (438), Expect = 9e-44
Identities = 93/301 (30%), Positives = 165/301 (53%), Gaps = 10/301 (3%)
Frame = +1
Query: 5 PVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKCMEQR 64
PV+D+GKL E+ +E + AC+ WGFF+ +NH ++ L++KV+ KE +E++
Sbjct: 403 PVIDLGKLLGEDA-TELEKLDLACKEWGFFQIINHGVNTSLIEKVKIGIKEFLSLPVEEK 579
Query: 65 FKEMVASKGLE-----CVQSEINDLDWESTFFLRHLP--SSNISEIPDLDEDYRKTMKEF 117
K +E V S+ L+W F + LP N P + + +R ++ +
Sbjct: 580 KKFWQTPNDMEGFGQMFVVSDDQKLEWADLFLITTLPLDERNPRLFPSIFQPFRDNLEIY 759
Query: 118 AEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSN---YPPCPKPDLIKGL 174
+++KL ++ + + L +E + ++F N T+ YPPCP+P+ + GL
Sbjct: 760 CSEVQKLCFTIISQMEKALKIESNEVTELF------NHITQAMRWNLYPPCPQPENVIGL 921
Query: 175 RAHTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVMH 234
H+D G + +L Q +++ GLQ+ KD QWI V P+P++ VIN+GD LE++TNG Y+S+ H
Sbjct: 922 NPHSDVGALTILLQANEIEGLQIRKDGQWIPVQPLPNAFVINIGDMLEIVTNGIYRSIEH 1101
Query: 235 RVIAQTDGARMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDYMKLYMGLKFQA 294
R ++ R+SIA+F+ P + ++SP +L+ + + + +Y+K Y K +
Sbjct: 1102RATVNSEKERISIAAFHRPHVNTILSPRPSLVTPERPAS-FKSIAVGEYLKAYFSRKLEG 1278
Query: 295 K 295
K
Sbjct: 1279K 1281
>TC89200 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (47%)
Length = 1323
Score = 171 bits (434), Expect = 3e-43
Identities = 90/304 (29%), Positives = 170/304 (55%), Gaps = 10/304 (3%)
Frame = +3
Query: 2 ENFPVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKCM 61
+ P++D+ KL +EE ++ AC+ WGFF+ +NH + + L++ +K +E +
Sbjct: 276 QQVPIIDLNKLLSEEDGTELQKFDLACKEWGFFQLINHGVDLSLVENFKKHVQEFFNLSA 455
Query: 62 EQRFKEMVASK-------GLECVQSEINDLDWESTFFLRHLPS--SNISEIPDLDEDYRK 112
E+ K++ K G V SE + L+W F++ LP+ N + P + + +R
Sbjct: 456 EE--KKIFGQKPGDMEGFGQMFVVSEEHKLEWADLFYIVTLPTYIRNPNLFPSIPQPFRD 629
Query: 113 TMKEFAEKLEKLAEELLDLLCENLGLEKGYLKKVFY-GSKGPNFGTKVSNYPPCPKPDLI 171
++ +A +++KL +++ + + L ++ L F GS+ +++ YPPCP+P+ +
Sbjct: 630 NLEMYAIEIKKLCVTIVEFMAKALKIQPNELLDFFEEGSQA----MRMNYYPPCPQPEQV 797
Query: 172 KGLRAHTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKS 231
GL H+D G + +L Q +++ GLQ+ KD WI + P+ ++ VIN+GD LE++TNG Y+S
Sbjct: 798 IGLNPHSDVGALTILLQLNEIEGLQVRKDGMWIPIKPLSNAFVINIGDMLEIMTNGIYRS 977
Query: 232 VMHRVIAQTDGARMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDYMKLYMGLK 291
+ HR + R+SIA+F + +A++ PA +L+ E+ ++ +D+ K Y K
Sbjct: 978 IEHRATINAEKERISIATFNSGRLNAILCPAPSLV-TPESPAVFNNVGVEDFFKAYFSRK 1154
Query: 292 FQAK 295
+ K
Sbjct: 1155LEGK 1166
>TC89553 weakly similar to SP|P24397|HY6H_HYONI Hyoscyamine 6-dioxygenase
(EC 1.14.11.11) (Hyoscyamine 6-beta- hydroxylase).
[Henbane], partial (49%)
Length = 1119
Score = 170 bits (431), Expect = 6e-43
Identities = 104/316 (32%), Positives = 164/316 (50%), Gaps = 9/316 (2%)
Frame = +1
Query: 2 ENFPVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKCM 61
+ P++D+G + T+ I A E +GFF+ +NH +S +LMD+ + +E +
Sbjct: 124 KTIPLIDLGG---HDHAHTILQILKASEEYGFFQVINHGVSKDLMDEAFNIFQEFHAMPP 294
Query: 62 EQRFKEMVAS-KGLEC---VQSEINDLD----WESTFFLRHLPSSNISEI-PDLDEDYRK 112
+++ E G+ C SE +D W+ T PS E P YR+
Sbjct: 295 KEKISECSRDPNGINCKIYASSENYKIDAVQYWKDTLTHPCPPSGEFMEFWPQKPPKYRE 474
Query: 113 TMKEFAEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIK 172
+ ++ ++L KL E+L++LCE LGL+ Y F G N ++PPCP+P L
Sbjct: 475 VVGKYTQELNKLGHEILEMLCEGLGLKLEY----FIGELSENPVILGHHFPPCPEPSLTL 642
Query: 173 GLRAHTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSV 232
GL H D I +L QD +V GLQ+LKDD+WI V P+P+++V+N+G L++ITNG+
Sbjct: 643 GLAKHRDPTIITILLQDQEVHGLQILKDDEWIPVEPIPNALVVNIGLILQIITNGRLVGA 822
Query: 233 MHRVIAQTDGARMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDYMKLYMGLKF 292
HRV+ + R S+A F P +I PA L+ + IY F ++ K KF
Sbjct: 823 EHRVVTNSKSVRTSVAYFIYPSFSRMIEPAKELV-DGNNPPIYKSMSFGEFRK-----KF 984
Query: 293 QAKEPRFEAMMKAMSS 308
K P+ E ++++ SS
Sbjct: 985 YDKGPKIEQVLQS*SS 1032
>TC78010 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (47%)
Length = 1321
Score = 169 bits (429), Expect = 1e-42
Identities = 88/292 (30%), Positives = 173/292 (59%), Gaps = 9/292 (3%)
Frame = +3
Query: 5 PVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKCMEQR 64
PV+D+ KL +EE + ++ + AC+ WGFF+ +NH ++ L++ +KL ++ + +E+
Sbjct: 222 PVIDLSKLLSEEDETELQKLDHACKEWGFFQLINHGVNPLLVENFKKLVQDFFNLPVEE- 398
Query: 65 FKEMVASK-------GLECVQSEINDLDWESTFFLRHLPS--SNISEIPDLDEDYRKTMK 115
K++++ K G V SE + L+W F + PS N P + + +R++++
Sbjct: 399 -KKILSQKPGNIEGFGQLFVVSEDHKLEWADLFHIITHPSYMRNPQLFPSIPQPFRESLE 575
Query: 116 EFAEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKGLR 175
++ L+KL +++ + + L ++K L + F+ G + +++ YPPCP+PD + GL
Sbjct: 576 MYSLVLKKLCVMIIEFMSKALKIQKNELLE-FFEEGGQSM--RMNYYPPCPQPDKVIGLN 746
Query: 176 AHTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVMHR 235
H+D + +L Q +++ GLQ+ KD WI + P+ ++ VIN+GD LE++TNG Y+S+ HR
Sbjct: 747 PHSDGTALTILLQLNEIEGLQIKKDGMWIPIKPLTNAFVINIGDMLEIMTNGIYRSIEHR 926
Query: 236 VIAQTDGARMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDYMKLY 287
++ R+SIA+F++ +A+++P +L+ +T ++ +D+ K Y
Sbjct: 927 ATINSEKERISIATFHSARLNAILAPVPSLI-TPKTPAVFNDISVEDFFKGY 1079
>TC78109 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (48%)
Length = 1314
Score = 118 bits (295), Expect(2) = 1e-42
Identities = 68/216 (31%), Positives = 117/216 (53%), Gaps = 12/216 (5%)
Frame = +3
Query: 1 MENFPVVDMGKLNTEE--RKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYK 58
+ PV+D+ KL + + ++ + AC+ WGFF+ +NH +S LM+ V+K +E +
Sbjct: 183 LPQLPVIDLSKLLSSHDLNEPELKKLHYACKEWGFFQLINHGVSESLMENVKKGAEEFFN 362
Query: 59 KCMEQRFK-----EMVASKGLECVQSEINDLDWESTFFLRHLPSSN-----ISEIPDLDE 108
ME++ K V G V SE LDW L LP S IP
Sbjct: 363 LPMEEKKKFGQTEGDVEGYGQAFVVSEEQKLDWADMLVLFTLPPHKRKPHLFSNIP---L 533
Query: 109 DYRKTMKEFAEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKP 168
+R ++ + EK+ LA +++DL+ +L ++ +++ F+G + T+++ YPPCP+P
Sbjct: 534 PFRVDLENYCEKMRTLAIQIMDLMAHSLAVDPMEIRE-FFGQATQS--TRMNYYPPCPQP 704
Query: 169 DLIKGLRAHTDAGGIILLFQDDKVSGLQLLKDDQWI 204
+ + GL +H+D GG+ +L Q +++ GLQ+ KD WI
Sbjct: 705 EFVIGLNSHSDGGGLTILLQGNEMDGLQIKKDGLWI 812
Score = 72.8 bits (177), Expect(2) = 1e-42
Identities = 35/100 (35%), Positives = 61/100 (61%)
Frame = +1
Query: 196 QLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVMHRVIAQTDGARMSIASFYNPGN 255
+L K D V P+P++ +INLGD LE++TNG ++S+ HR ++ R SIASFY+P
Sbjct: 787 KLKKMDCGFHVKPLPNAFIINLGDMLEMMTNGIFRSIEHRATVNSEKERFSIASFYSPAF 966
Query: 256 DAVISPASTLLKEDETSEIYPKFIFDDYMKLYMGLKFQAK 295
+ ++SPA +++ + T ++ + +Y K Y+ + K
Sbjct: 967 NTILSPAPSIVTPN-TPAVFKRISAGEYFKGYLAQELCGK 1083
>TC87461 weakly similar to GP|2688828|gb|AAB88878.1|
ethylene-forming-enzyme-like dioxygenase {Prunus
armeniaca}, partial (62%)
Length = 993
Score = 169 bits (428), Expect = 1e-42
Identities = 89/273 (32%), Positives = 157/273 (56%), Gaps = 9/273 (3%)
Frame = +2
Query: 3 NFPVVDMGKL-NTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKCM 61
+ PVVD+GKL + + + + A +WG F+ +NH ++ ++KV +++K+ ++
Sbjct: 170 HIPVVDIGKLISPSTSQQELHKLHSALSSWGLFQAINHGMTSLTLNKVREISKQFFELSK 349
Query: 62 EQRFKEMVASKGLE-----CVQSEINDLDWESTFFLRHLPSS--NISEIPDLDEDYRKTM 114
E++ K G+E + SE LDW +L+ P N P D+R T+
Sbjct: 350 EEKQKYAREPNGIEGYGNDVILSENQKLDWTDRVYLKVHPEHQRNFKLFPQKPNDFRNTI 529
Query: 115 KEFAEKLEKLAEELLDLLCENLGLEKG-YLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKG 173
+++ + L +L E +L + ++L LE+ +LK+ G + F +++ YPPCP PD + G
Sbjct: 530 EQYTQSLRQLYEIILRAVAKSLNLEEDCFLKEC--GERDTMF-MRINYYPPCPMPDHVLG 700
Query: 174 LRAHTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVM 233
++ H D I L QD +V GLQ+LKD+QW VP +P ++VIN+GDQ+E+++NG ++S +
Sbjct: 701 VKPHADGSSITFLLQDKEVEGLQVLKDNQWFKVPIIPDALVINVGDQIEIMSNGIFQSPV 880
Query: 234 HRVIAQTDGARMSIASFYNPGNDAVISPASTLL 266
HRV+ + R+++A F P ++ I P L+
Sbjct: 881 HRVVINEEKERLTVAMFCIPDSEQEIKPVDKLV 979
>TC86474 weakly similar to GP|6016680|gb|AAF01507.1| putative
leucoanthocyanidin dioxygenase {Arabidopsis thaliana},
partial (20%)
Length = 1395
Score = 165 bits (418), Expect = 2e-41
Identities = 91/298 (30%), Positives = 165/298 (54%), Gaps = 7/298 (2%)
Frame = +1
Query: 5 PVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKCMEQR 64
PV+D G L+ + +++ AC+ WGFF+ VNH + I+LM +++ + E + +E++
Sbjct: 238 PVIDFGLLSHGNKNELLKL-DIACKEWGFFQIVNHGMEIDLMQRLKDVVAEFFDLSIEEK 414
Query: 65 FK-----EMVASKGLECVQSEINDLDWESTFFLRHLPSS--NISEIPDLDEDYRKTMKEF 117
K + + G V SE LDW L P+ P+ E + T++ +
Sbjct: 415 DKYAMPPDDIQGYGHTSVVSEKQILDWCDQLILLVYPTRFRKPQFWPETPEKLKDTIEAY 594
Query: 118 AEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKGLRAH 177
+ +++++ EEL++ L GLE+ L + K G +V+ YPPC P+ + GL H
Sbjct: 595 SSEIKRVGEELINSLSLIFGLEEHVLLGLH---KEVLQGLRVNYYPPCNTPEQVIGLTPH 765
Query: 178 TDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVMHRVI 237
+DA + ++ QDD V+GL++ W+ + P+P+++V+NLGD +EV++NGKYKSV HR +
Sbjct: 766 SDASTVTIVMQDDDVTGLEVRYKGNWVPINPIPNALVVNLGDVIEVLSNGKYKSVEHRAM 945
Query: 238 AQTDGARMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDYMKLYMGLKFQAK 295
+ R S SF P +DA + P ++ +D+ ++Y + + +Y++ + K + K
Sbjct: 946 TNKNKRRTSFVSFLFPRDDAELGPFDHMI-DDQNPKMYKEITYGEYLRHTLNRKLEGK 1116
>TC77355 weakly similar to PIR|T05552|T05552 SRG1 protein-related protein
F24A6.150 - Arabidopsis thaliana, partial (44%)
Length = 1359
Score = 162 bits (410), Expect = 2e-40
Identities = 92/298 (30%), Positives = 160/298 (52%), Gaps = 7/298 (2%)
Frame = +1
Query: 5 PVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKCMEQR 64
PV+D +L+ + +++ + AC+ WGFF+ VNH + +L+ ++ + E ++ ME++
Sbjct: 202 PVIDFTQLSNGNMEELLKL-EIACKEWGFFQMVNHGVEKDLIQRMRDASNEFFELPMEEK 378
Query: 65 FKEM-----VASKGLECVQSEINDLDWESTFFLRHLPSS--NISEIPDLDEDYRKTMKEF 117
K + G V SE LDW L P + P +++ ++ +
Sbjct: 379 EKYAMLPNDIQGYGQNYVVSEEQTLDWSDVLVLIIYPDRYRKLQFWPKTSHGFKEIIEAY 558
Query: 118 AEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKGLRAH 177
+ ++ ++ EELL L +GLEK L ++ K G +V+ YPPC P+ + GL H
Sbjct: 559 SSEVRRVGEELLSFLSIIMGLEKHALAELH---KELIQGLRVNYYPPCNNPEQVLGLSPH 729
Query: 178 TDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVMHRVI 237
+D I LL QDD V GL++ W+ V P+ ++VIN+GD +E+++NGKYKSV HRV+
Sbjct: 730 SDTTTITLLIQDDDVLGLEIRNKGNWVPVKPISDALVINVGDVIEILSNGKYKSVEHRVM 909
Query: 238 AQTDGARMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDYMKLYMGLKFQAK 295
+ R+S ASF P +D + P ++ + + ++Y K + DY++ + K + K
Sbjct: 910 TNQNKRRISYASFLFPRDDVEVEPFDHMI-DAQNPKMYQKVKYGDYLRHSLKRKMERK 1080
>TC82784 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (59%)
Length = 1206
Score = 162 bits (409), Expect = 2e-40
Identities = 93/300 (31%), Positives = 167/300 (55%), Gaps = 9/300 (3%)
Frame = +3
Query: 5 PVVDMGKL-NTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKCMEQ 63
PV+DM KL + E + + AC++WGFF+ VNH +S L++KV+ + + M +
Sbjct: 132 PVIDMKKLLSLEYGSLELSKLHLACKDWGFFQLVNHDVSSSLLEKVKMEIMDFFNLPMSE 311
Query: 64 RFK-----EMVASKGLECVQSEINDLDWESTFFLRHLPSSNISE--IPDLDEDYRKTMKE 116
+ K + + G V SE LDW FF+ LP P L R T++
Sbjct: 312 KKKFWQTPQHMEGFGQAFVLSEEQKLDWADLFFMTTLPKHLRMPHLFPQLPLPLRDTLEL 491
Query: 117 FAEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKGLRA 176
++++++ LA +L + ++L +E+ ++++F G + + YPPCP+P+ + GL
Sbjct: 492 YSQEIKNLAMVILGHIEKSLKMEEMEIRELF--EDGIQM-MRTNYYPPCPQPEKVIGLTN 662
Query: 177 HTDAGGIILLFQ-DDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVMHR 235
H+D G+ +L Q ++ GLQ+ K+ W+ V P+P++ ++N+GD LE+ITNG Y+S+ HR
Sbjct: 663 HSDPVGLTILLQLNESRKGLQIRKNCMWVPVKPLPNAFIVNIGDMLEIITNGIYRSIEHR 842
Query: 236 VIAQTDGARMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDYMKLYMGLKFQAK 295
I ++ R+SIA+FY+ + +++ P +L+ E +T + K ++Y K + K
Sbjct: 843 AIVNSEKERLSIATFYSSRHGSILGPVKSLITE-QTPARFKKVGVEEYFTNLFARKLEGK 1019
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.136 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,153,300
Number of Sequences: 36976
Number of extensions: 121169
Number of successful extensions: 883
Number of sequences better than 10.0: 176
Number of HSP's better than 10.0 without gapping: 750
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 754
length of query: 317
length of database: 9,014,727
effective HSP length: 96
effective length of query: 221
effective length of database: 5,465,031
effective search space: 1207771851
effective search space used: 1207771851
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC122163.7


