

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122163.6 + phase: 0
(459 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
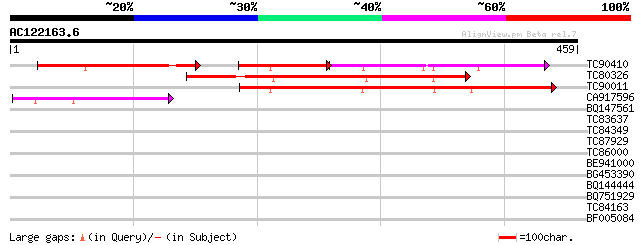
Score E
Sequences producing significant alignments: (bits) Value
TC90410 similar to PIR|T04638|T04638 hypothetical protein F10N7.... 140 1e-76
TC80326 similar to GP|1732517|gb|AAB38779.1| putative cytoskelet... 246 1e-65
TC90011 similar to PIR|T04638|T04638 hypothetical protein F10N7.... 216 1e-56
CA917596 similar to PIR|T01449|T01 cytoskeletal protein homolog ... 117 1e-26
BQ147561 similar to PIR|T13758|T137 NADH dehydrogenase (ubiquino... 35 0.046
TC83637 similar to GP|9279731|dbj|BAB01321.1 disease resistance ... 31 1.1
TC84349 similar to PIR|D86390|D86390 hypothetical protein AAF985... 29 3.3
TC87929 similar to GP|12006424|gb|AAG44852.1 Ku70-like protein {... 29 3.3
TC86000 phosphoinositide-specific phospholipase C [Medicago trun... 29 4.3
BE941000 28 5.7
BG453390 similar to GP|22202666|dbj putative male sterility 1 pr... 28 5.7
BQ144444 GP|18376070|em related to tpa inducible protein {Neuros... 28 7.4
BQ751929 28 7.4
TC84163 similar to GP|13365608|dbj|BAB39154. avicelase 2 {Humico... 28 9.7
BF005084 weakly similar to GP|9758958|dbj| gene_id:MRB17.16~unkn... 28 9.7
>TC90410 similar to PIR|T04638|T04638 hypothetical protein F10N7.120 -
Arabidopsis thaliana, partial (23%)
Length = 1983
Score = 140 bits (354), Expect(3) = 1e-76
Identities = 86/206 (41%), Positives = 120/206 (57%), Gaps = 28/206 (13%)
Frame = +3
Query: 260 GNVHMCAARKFVRMDENENEVLVMNE---SEFDFVQEKYYLAREKYEQAVVIKPDFYEGL 316
GNVHM ARK V + E ++ M E S +++ Q++Y A EK+E A+ IK DFYEG
Sbjct: 732 GNVHMSRARKKVYLTEEDSSKEHMCEQIKSSYEWAQKEYAKAGEKFEAAIKIKSDFYEGF 911
Query: 317 LAIGQQQFELAKLNWSF--GIANKMDL----GKETLRLFDVAEEKMTAANDAWENLEKGK 370
LA+GQQ+FE AKL+ F ++N +DL E L L++ AEE M WE ++ +
Sbjct: 912 LALGQQRFEQAKLSLVFMHSVSN-IDLPTWPSTEVLHLYNNAEENMEKGMLIWEESQEQQ 1088
Query: 371 LGEQGSVG-------------------MRSQIHLFWGNMLFERSQVEFKLGMSDWKKKLD 411
L E + G MRSQI+L WG ML+ERS V+FKLG+ W + L+
Sbjct: 1089LNETSTTGLDGLFENMSSDETAVLAANMRSQINLLWGTMLYERSLVKFKLGLPVWHESLE 1268
Query: 412 ASVERFKIAGASEADVSGILKKHCFN 437
++E+F+ AGAS DV+ +LK HC N
Sbjct: 1269TAIEKFEHAGASPTDVAVMLKNHCSN 1346
Score = 102 bits (254), Expect(3) = 1e-76
Identities = 56/137 (40%), Positives = 84/137 (60%), Gaps = 5/137 (3%)
Frame = +1
Query: 23 NIAFEPHPSPKKSDQISRRLREPKLGPTNEHLKENQKE-----DVKIAWRQLKLVYDDDI 77
N A + H KKS++ + + K+ E +KE +E V + + KLV+ DDI
Sbjct: 22 NKAEDDHLKEKKSNKSKNKAAKEKIDKQKEDVKEVNEEKSNGRSVDVPKKTAKLVFGDDI 201
Query: 78 RLAQMPINCSFRLLRDIVKEKFPISRSVLIKYKDNDDDLVTITSTEELRFAESCVYKTDS 137
R AQ+PINCS LR+++ ++FP +VL+KY+D + DL+TITS EEL++AE T S
Sbjct: 202 RWAQLPINCSLFQLREVICDRFPSLGAVLVKYRDQEGDLITITSDEELKWAE-----TGS 366
Query: 138 VEILKLYIVEVSPEHEP 154
++LYIVE +P H+P
Sbjct: 367 QGSIRLYIVEANPNHDP 417
Score = 83.6 bits (205), Expect(3) = 1e-76
Identities = 40/77 (51%), Positives = 58/77 (74%), Gaps = 2/77 (2%)
Frame = +2
Query: 186 KVVENLEIDDWLYEFAQLFRSRVG--TDKYIDFHDLGTEFCSDALEETVTSDEAQDLLDK 243
K++ + ++DW+ +FA LF++ VG +D+Y+DFH+LG + S+A+EETVTS+EAQ L D
Sbjct: 503 KLLASSCVEDWIIKFADLFKNHVGFESDRYLDFHELGMKLYSEAVEETVTSEEAQGLFDM 682
Query: 244 AEFKFQEVAALAFFNWG 260
A KFQE+ ALA FN G
Sbjct: 683 AGGKFQEMTALALFNCG 733
>TC80326 similar to GP|1732517|gb|AAB38779.1| putative cytoskeletal protein
{Arabidopsis thaliana}, partial (29%)
Length = 833
Score = 246 bits (629), Expect = 1e-65
Identities = 132/238 (55%), Positives = 168/238 (70%), Gaps = 8/238 (3%)
Frame = +1
Query: 144 YIVEVSPEHEPPLLKEEKEEENNEKQKPLDCVLDEKMCTECNKVVENLEIDDWLYEFAQL 203
++ EV+ E P + +K + K + C E +E+DDWL+EFAQL
Sbjct: 67 FVPEVTEVTEVPDTEVDKIITKKDVSKEKPGATGDNECKE-------VEMDDWLFEFAQL 225
Query: 204 FRSRVGTDK--YIDFHDLGTEFCSDALEETVTSDEAQDLLDKAEFKFQEVAALAFFNWGN 261
FRS VG D +ID H+LG E CS+ALEETVTS+EAQDL DKA KFQEVAALAFFNWGN
Sbjct: 226 FRSHVGIDPDAHIDLHELGMELCSEALEETVTSEEAQDLFDKAASKFQEVAALAFFNWGN 405
Query: 262 VHMCAARKFVRMDENENEVLVMNESE--FDFVQEKYYLAREKYEQAVVIKPDFYEGLLAI 319
VHMCAARK + +DE+ + +V + + +D+V+EKY LAREKYE+A++IKPDFYEGLLA+
Sbjct: 406 VHMCAARKRIPLDESAGKDVVAEQLQVAYDWVKEKYSLAREKYEEALLIKPDFYEGLLAL 585
Query: 320 GQQQFELAKLNWSFGIANKMDL----GKETLRLFDVAEEKMTAANDAWENLEKGKLGE 373
GQQQFE+AKL+WSF IA K+DL ETL+LF+ AEEKM +A D WE LE+ + E
Sbjct: 586 GQQQFEMAKLHWSFAIAKKIDLSTWDSTETLQLFNSAEEKMKSATDMWEKLEEQRAKE 759
>TC90011 similar to PIR|T04638|T04638 hypothetical protein F10N7.120 -
Arabidopsis thaliana, partial (33%)
Length = 1324
Score = 216 bits (551), Expect = 1e-56
Identities = 116/293 (39%), Positives = 179/293 (60%), Gaps = 37/293 (12%)
Frame = +1
Query: 187 VVENLEIDDWLYEFAQLFRSRVG--TDKYIDFHDLGTEFCSDALEETVTSDEAQDLLDKA 244
V+ L +++WL +FA+LF++ VG +D Y+D H++G + S+A+E++VT+D AQ+L D A
Sbjct: 4 VIG*LPVEEWLVQFARLFKNHVGFDSDSYLDIHEVGMKIYSEAMEDSVTNDGAQELFDIA 183
Query: 245 EFKFQEVAALAFFNWGNVHMCAARKFVRMDENENEVLVMN--ESEFDFVQEKYYLAREKY 302
KFQE+AALA FNWGNVH+ AR+ V E+ + ++ +++ ++Y A +Y
Sbjct: 184 ADKFQEMAALALFNWGNVHLSKARRRVPFQEDGTREASLEHLKAGYEWATKEYKEAEMRY 363
Query: 303 EQAVVIKPDFYEGLLAIGQQQFELAKLNWSFGIANKMDLG----KETLRLFDVAEEKMTA 358
E++V IKPDFYEGLLA+G QQFE AKL W + IAN ++ +E L+L++ AE+ M
Sbjct: 364 EESVKIKPDFYEGLLALGYQQFEQAKLCWCYLIANNKNIEVGPFEEVLQLYNKAEDSMEK 543
Query: 359 ANDAWENLEKGKLG-----------------------------EQGSVGMRSQIHLFWGN 389
WE +E+ +L E+ + MR QI+L WG
Sbjct: 544 GMLMWEEIEEQRLNGLSKLDKYNAELEKMGLHGLFRDISSEEAEEQASNMRIQIYLLWGT 723
Query: 390 MLFERSQVEFKLGMSDWKKKLDASVERFKIAGASEADVSGILKKHCFNGNARD 442
+L+ERS VEFKLG+ W++ L+ +VE+F++AGAS D+ ++K HC N A +
Sbjct: 724 LLYERSVVEFKLGLPTWEECLEVAVEKFELAGASTTDIGVMIKNHCSNETAME 882
>CA917596 similar to PIR|T01449|T01 cytoskeletal protein homolog F24O1.11 -
Arabidopsis thaliana, partial (16%)
Length = 616
Score = 117 bits (292), Expect = 1e-26
Identities = 68/139 (48%), Positives = 84/139 (59%), Gaps = 9/139 (6%)
Frame = +1
Query: 3 RSGSRRKKGNVSQPNSP--SSSNIAFEPHPSPKKSDQISRRLREP-KLGPT------NEH 53
+ G G+V PN+ S N+ + Q+ + + +P GP NE
Sbjct: 178 KKGVNSAVGSVVSPNNKVDKSQNVLLPTENGLENKTQMPKVVLKPFNNGPVVQSNSKNES 357
Query: 54 LKENQKEDVKIAWRQLKLVYDDDIRLAQMPINCSFRLLRDIVKEKFPISRSVLIKYKDND 113
K+ +V I WR LKLVYD DIRLAQMP NCSFR+LRD+V ++FP S SVLIKYKD D
Sbjct: 358 QKDRNLSEVAIRWRPLKLVYDHDIRLAQMPANCSFRVLRDVVSKRFPSSNSVLIKYKDCD 537
Query: 114 DDLVTITSTEELRFAESCV 132
DLVTITST+ELR AES V
Sbjct: 538 GDLVTITSTDELRLAESFV 594
>BQ147561 similar to PIR|T13758|T137 NADH dehydrogenase (ubiquinone) (EC
1.6.5.3) chain 5 - Tipularia discolor chloroplast
(fragment), partial (4%)
Length = 600
Score = 35.4 bits (80), Expect = 0.046
Identities = 37/161 (22%), Positives = 71/161 (43%)
Frame = +2
Query: 30 PSPKKSDQISRRLREPKLGPTNEHLKENQKEDVKIAWRQLKLVYDDDIRLAQMPINCSFR 89
P PK + + P P E +KE+ E A +Q +L+ Q +N
Sbjct: 89 PKPKVTKDGGVDXKAPXPEPEPEQVKEDSVETKVEAEKQEQLL--------QSSLNLDVN 244
Query: 90 LLRDIVKEKFPISRSVLIKYKDNDDDLVTITSTEELRFAESCVYKTDSVEILKLYIVEVS 149
+ D ++ RS+ + + ++D T E+ + E+ K +++E K +E +
Sbjct: 245 EIVD--DDQANKRRSLSLLFNKENEDAKDSTENEKNKAEETV--KEETLETEKP--LEDA 406
Query: 150 PEHEPPLLKEEKEEENNEKQKPLDCVLDEKMCTECNKVVEN 190
+EP L K +E +NNE +C + E+ + NK+ ++
Sbjct: 407 KSNEPTLEKPSQETKNNEPSTEQECAVVEEKPSLQNKITDS 529
>TC83637 similar to GP|9279731|dbj|BAB01321.1 disease resistance protein
RPP1-WsB {Arabidopsis thaliana}, partial (2%)
Length = 1045
Score = 30.8 bits (68), Expect = 1.1
Identities = 30/109 (27%), Positives = 45/109 (40%), Gaps = 4/109 (3%)
Frame = +3
Query: 52 EHLKENQKEDVKIAWRQLKLVYDDDIRLAQMPINCSFRLLRDIVKEKFPISRS----VLI 107
E KEN+KE VK W L L D+ L + +N L++ ++ + +
Sbjct: 672 EQFKENKKE-VKF-WNCLNL---DERSLINIGLNLQINLMKFAYQDLSTVEHDDYVETYV 836
Query: 108 KYKDNDDDLVTITSTEELRFAESCVYKTDSVEILKLYIVEVSPEHEPPL 156
YKDN D + + YKT + IV++SP H PL
Sbjct: 837 DYKDNFDSYQALYVYSGSSVPDWFEYKTTNETTNDDMIVDLSPLHLSPL 983
>TC84349 similar to PIR|D86390|D86390 hypothetical protein AAF98584.1
[imported] - Arabidopsis thaliana, partial (15%)
Length = 685
Score = 29.3 bits (64), Expect = 3.3
Identities = 15/50 (30%), Positives = 26/50 (52%)
Frame = +2
Query: 15 QPNSPSSSNIAFEPHPSPKKSDQISRRLREPKLGPTNEHLKENQKEDVKI 64
QPN+ +++N + H SP++ I +R P ++E QK D+ I
Sbjct: 248 QPNANANANRNNQTHSSPRRQKIIEQRKSLPIASVEKRLIEEVQKNDILI 397
>TC87929 similar to GP|12006424|gb|AAG44852.1 Ku70-like protein {Arabidopsis
thaliana}, partial (36%)
Length = 1279
Score = 29.3 bits (64), Expect = 3.3
Identities = 20/85 (23%), Positives = 35/85 (40%)
Frame = +1
Query: 109 YKDNDDDLVTITSTEELRFAESCVYKTDSVEILKLYIVEVSPEHEPPLLKEEKEEENNEK 168
++D++DD S + DS + +Y+V+ SP E EE+ +
Sbjct: 115 FRDDEDDPDAQNSFQN---------NNDSNKEFVVYLVDASPNMFITTSSSESEEQESHF 267
Query: 169 QKPLDCVLDEKMCTECNKVVENLEI 193
Q L C+ D NK ++ + I
Sbjct: 268 QIALSCISDSLKAMIINKSLDQIAI 342
>TC86000 phosphoinositide-specific phospholipase C [Medicago truncatula]
Length = 2160
Score = 28.9 bits (63), Expect = 4.3
Identities = 31/103 (30%), Positives = 40/103 (38%), Gaps = 3/103 (2%)
Frame = +3
Query: 142 KLYIVEVSPEHEPPLLKEEKEEENNEKQKPLDCVLDEKMCTECNKVVENLEIDDWLYEFA 201
K I+ P E KEEKE+E ++K KPL DE + W E
Sbjct: 876 KRIIISTKPPKEYLEAKEEKEKEESQKGKPLG---DE---------------EAWGKEVP 1001
Query: 202 QLFRSRVGTDKY---IDFHDLGTEFCSDALEETVTSDEAQDLL 241
L + K ID DL E SD TSD+ + L+
Sbjct: 1002SLRGGTIADYKQNSGIDEDDLKEEEDSDEASRQNTSDDYRRLI 1130
>BE941000
Length = 440
Score = 28.5 bits (62), Expect = 5.7
Identities = 12/31 (38%), Positives = 17/31 (54%)
Frame = +2
Query: 23 NIAFEPHPSPKKSDQISRRLREPKLGPTNEH 53
N F P+P+P + S R + +L PT EH
Sbjct: 290 NAGFAPNPNPLNLIKSSNRFDQNQLNPTQEH 382
>BG453390 similar to GP|22202666|dbj putative male sterility 1 protein {Oryza
sativa (japonica cultivar-group)}, partial (5%)
Length = 655
Score = 28.5 bits (62), Expect = 5.7
Identities = 9/23 (39%), Positives = 14/23 (60%)
Frame = +3
Query: 176 LDEKMCTECNKVVENLEIDDWLY 198
L E C CN V + +++DW+Y
Sbjct: 504 LSESKCKSCNHVTTSDDVEDWVY 572
>BQ144444 GP|18376070|em related to tpa inducible protein {Neurospora
crassa}, partial (2%)
Length = 1256
Score = 28.1 bits (61), Expect = 7.4
Identities = 19/65 (29%), Positives = 24/65 (36%), Gaps = 21/65 (32%)
Frame = -2
Query: 7 RRKKGNVSQPNSPSSSNIAFEPHPSPKKSD---------------------QISRRLREP 45
RRK+ ++P +P PHP P KS SRR R+P
Sbjct: 622 RRKEKKAARPRTPPPPPPPSVPHPHPNKSHIYTTARAYTPIYHALCHPYPRYRSRRTRQP 443
Query: 46 KLGPT 50
GPT
Sbjct: 442 PTGPT 428
>BQ751929
Length = 771
Score = 28.1 bits (61), Expect = 7.4
Identities = 25/115 (21%), Positives = 54/115 (46%), Gaps = 23/115 (20%)
Frame = -2
Query: 39 SRRLREPKLGPTNEHLKENQKEDVKIAWRQLKLVYDDDI-----RLAQMPIN-----CSF 88
SR+ R+P +G + + E+ D+ + + K ++D+DI RL Q + C
Sbjct: 755 SRQQRKPTMGENHGGIPEHGIHDIHVELPRAKRLFDEDIVRGRVRLEQHDVGTENLLCGR 576
Query: 89 RLL-------RD------IVKEKFPISRSVLIKYKDNDDDLVTITSTEELRFAES 130
R+L RD ++K+ + + + ++ + D+D + + E ++F E+
Sbjct: 575 RVLFIQPVRPRDSNSGLGLLKQVIHVVQDLRVRVE--DEDALVVGQLEGVKFVEA 417
>TC84163 similar to GP|13365608|dbj|BAB39154. avicelase 2 {Humicola
insolens}, partial (3%)
Length = 513
Score = 27.7 bits (60), Expect = 9.7
Identities = 8/22 (36%), Positives = 17/22 (76%)
Frame = -1
Query: 268 RKFVRMDENENEVLVMNESEFD 289
+ ++ +D NEN++ ++NE E+D
Sbjct: 405 KNYIHLDFNENQISLINEDEYD 340
>BF005084 weakly similar to GP|9758958|dbj| gene_id:MRB17.16~unknown protein
{Arabidopsis thaliana}, partial (38%)
Length = 603
Score = 27.7 bits (60), Expect = 9.7
Identities = 12/32 (37%), Positives = 18/32 (55%)
Frame = -1
Query: 2 RRSGSRRKKGNVSQPNSPSSSNIAFEPHPSPK 33
+ SGS +GN S+ + +NI + PSPK
Sbjct: 246 KTSGSNSHRGNASEGSKNFPNNIGLKSDPSPK 151
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.134 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,993,533
Number of Sequences: 36976
Number of extensions: 161364
Number of successful extensions: 959
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 940
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 947
length of query: 459
length of database: 9,014,727
effective HSP length: 99
effective length of query: 360
effective length of database: 5,354,103
effective search space: 1927477080
effective search space used: 1927477080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC122163.6


