

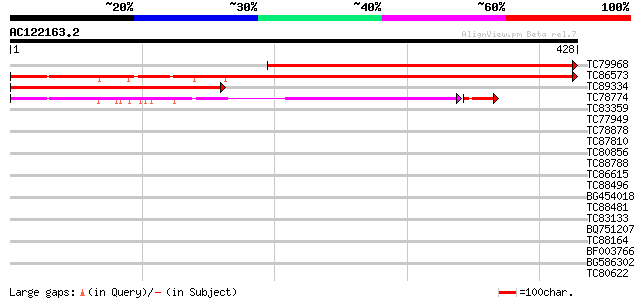
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122163.2 + phase: 0
(428 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC79968 similar to GP|22655310|gb|AAM98245.1 unknown protein {Ar... 483 e-137
TC86573 similar to GP|15292907|gb|AAK92824.1 unknown protein {Ar... 444 e-125
TC89334 similar to GP|22655310|gb|AAM98245.1 unknown protein {Ar... 323 1e-88
TC78774 similar to GP|15292907|gb|AAK92824.1 unknown protein {Ar... 271 1e-76
TC83359 weakly similar to PIR|G86385|G86385 hypothetical protein... 39 0.005
TC77949 weakly similar to GP|11762218|gb|AAG40387.1 AT4g27520 {A... 38 0.007
TC78878 similar to GP|4519792|dbj|BAA75744.1 Asp1 {Arabidopsis t... 33 0.21
TC87810 similar to GP|21592752|gb|AAM64701.1 unknown {Arabidopsi... 33 0.28
TC80856 similar to GP|15706274|emb|CAC69997. HMG I/Y like protei... 32 0.36
TC88788 weakly similar to SP|P05790|FBOH_BOMMO Fibroin heavy cha... 32 0.47
TC86615 weakly similar to GP|8778409|gb|AAF79417.1| F16A14.5 {Ar... 31 1.0
TC88496 similar to GP|1079652|gb|AAD09209.1| late embryogenesis ... 30 1.4
BG454018 similar to GP|1079652|gb| late embryogenesis abundant p... 30 1.8
TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclea... 30 1.8
TC83133 similar to PIR|T06033|T06033 hypothetical protein T28I19... 30 2.3
BQ751207 weakly similar to GP|604427|gb|AA ACOB protein {Emerice... 30 2.3
TC88164 GP|3763933|gb|AAC64313.1| unknown protein {Arabidopsis t... 29 3.0
BF003766 similar to PIR|S56032|S560 probable membrane protein YJ... 29 3.0
BG586302 29 4.0
TC80622 homologue to PIR|T05792|T05792 hypothetical protein M7J2... 29 4.0
>TC79968 similar to GP|22655310|gb|AAM98245.1 unknown protein {Arabidopsis
thaliana}, partial (49%)
Length = 1230
Score = 483 bits (1244), Expect = e-137
Identities = 234/234 (100%), Positives = 234/234 (100%)
Frame = +2
Query: 195 LGGVIVPVLVLIIWNCVLGRKGLLGFVKRYPDAELRGAIDGQYVKVTGVVTCGSIPLESS 254
LGGVIVPVLVLIIWNCVLGRKGLLGFVKRYPDAELRGAIDGQYVKVTGVVTCGSIPLESS
Sbjct: 2 LGGVIVPVLVLIIWNCVLGRKGLLGFVKRYPDAELRGAIDGQYVKVTGVVTCGSIPLESS 181
Query: 255 YQRIPRCVYVSSELYEYKGWGGKSAHPKHRCFTWGSRYSEKYIADFYISDFQTGLRALVK 314
YQRIPRCVYVSSELYEYKGWGGKSAHPKHRCFTWGSRYSEKYIADFYISDFQTGLRALVK
Sbjct: 182 YQRIPRCVYVSSELYEYKGWGGKSAHPKHRCFTWGSRYSEKYIADFYISDFQTGLRALVK 361
Query: 315 AGYGNKVAPFVKPTTVVDVTKENRELSPNFLGWLADRKLSTDDRIMRLKEGHIKEGSTVS 374
AGYGNKVAPFVKPTTVVDVTKENRELSPNFLGWLADRKLSTDDRIMRLKEGHIKEGSTVS
Sbjct: 362 AGYGNKVAPFVKPTTVVDVTKENRELSPNFLGWLADRKLSTDDRIMRLKEGHIKEGSTVS 541
Query: 375 VMGVVRRHENVLMIVPPTEPVSTGCQWMRCLLPTGVEGLIITCEDNQNADVIAV 428
VMGVVRRHENVLMIVPPTEPVSTGCQWMRCLLPTGVEGLIITCEDNQNADVIAV
Sbjct: 542 VMGVVRRHENVLMIVPPTEPVSTGCQWMRCLLPTGVEGLIITCEDNQNADVIAV 703
>TC86573 similar to GP|15292907|gb|AAK92824.1 unknown protein {Arabidopsis
thaliana}, partial (92%)
Length = 2060
Score = 444 bits (1142), Expect = e-125
Identities = 236/483 (48%), Positives = 315/483 (64%), Gaps = 55/483 (11%)
Frame = +3
Query: 1 MGTRIPSHQLSSGLYVSGRPEQPKERQPPTMASRSVPYTGGDPKKSGELGKMLDIPVLDP 60
MG+R PSHQLS+GLYVSGRPEQPKER P TM+S ++PYTGGD KKSGELGKM DIP+
Sbjct: 255 MGSRFPSHQLSNGLYVSGRPEQPKERTP-TMSSVAMPYTGGDIKKSGELGKMFDIPMDGS 431
Query: 61 KSHPSS--------------------------------------SSSQLSTGPARSRPNS 82
KS S S++ +ST + R NS
Sbjct: 432 KSRKSGPLNNAPSRTGSFAGAASHSGPIMQNAAARSAYVTSGNLSAAGMSTSASMKRTNS 611
Query: 83 GQVGKN----------ITGSGTLSRKSTGSGPIALQPTGLITSGPVGSGPVGASRR-SGQ 131
G + K+ +G GT R+++G P L TGLITSGP+ S GA R+ SG
Sbjct: 612 GPLNKHGEPVKKSSGPQSGGGT--RQNSGPIPPVLPTTGLITSGPLNSS--GAPRKVSGP 779
Query: 132 LEQSGSM----GKAVYGSAVTSLGEEVKVGFRVS--RSVVWVFMVVVAMCLLVGVFLMVA 185
LE GSM V+ AVT+L + + FR + + ++W +++ M + G F++ A
Sbjct: 780 LESLGSMKLNSASVVHNPAVTTLSVDDEYSFRKNFPKPILWSVILIFVMGFIAGGFILGA 959
Query: 186 VKKNVILFALGGVIVPVLVLIIWNCVLGRKGLLGFVKRYPDAELRGAIDGQYVKVTGVVT 245
V ++L + + V L WN GRK ++GF+ +YPDAELR A +GQ+VKV+GVVT
Sbjct: 960 VHNAILLIVVVILFAAVAALFTWNSCRGRKAIVGFISQYPDAELRTAKNGQFVKVSGVVT 1139
Query: 246 CGSIPLESSYQRIPRCVYVSSELYEYKGWGGKSAHPKHRCFTWGSRYSEKYIADFYISDF 305
CG++PLESS+Q++PRCVY S+ LYEY+GW K+A+ KHR FTWG R +E+++ DFYISDF
Sbjct: 1140CGNVPLESSFQKVPRCVYTSTSLYEYRGWDSKAANSKHRRFTWGLRSTERHVVDFYISDF 1319
Query: 306 QTGLRALVKAGYGNKVAPFVKPTTVVDVTKENRELSPNFLGWLADRKLSTDDRIMRLKEG 365
Q+GLRALVK G+G +V P+V + V+DV EN+++SP+FL WL R LS+DDRIM+LKEG
Sbjct: 1320QSGLRALVKTGFGARVTPYVDDSVVIDVNPENKDMSPDFLRWLGKRNLSSDDRIMQLKEG 1499
Query: 366 HIKEGSTVSVMGVVRRHENVLMIVPPTEPVSTGCQWMRCLLPTGVEGLIITCEDNQNADV 425
+IKEGSTVSVMGVV R++NVLMIVPP EP++TGCQW +C+ P +EG+++ CED DV
Sbjct: 1500YIKEGSTVSVMGVVHRNDNVLMIVPPPEPLTTGCQWAKCIFPASLEGIVLRCEDTSKIDV 1679
Query: 426 IAV 428
I V
Sbjct: 1680IPV 1688
>TC89334 similar to GP|22655310|gb|AAM98245.1 unknown protein {Arabidopsis
thaliana}, partial (17%)
Length = 624
Score = 323 bits (827), Expect = 1e-88
Identities = 163/163 (100%), Positives = 163/163 (100%)
Frame = +1
Query: 1 MGTRIPSHQLSSGLYVSGRPEQPKERQPPTMASRSVPYTGGDPKKSGELGKMLDIPVLDP 60
MGTRIPSHQLSSGLYVSGRPEQPKERQPPTMASRSVPYTGGDPKKSGELGKMLDIPVLDP
Sbjct: 121 MGTRIPSHQLSSGLYVSGRPEQPKERQPPTMASRSVPYTGGDPKKSGELGKMLDIPVLDP 300
Query: 61 KSHPSSSSSQLSTGPARSRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLITSGPVGS 120
KSHPSSSSSQLSTGPARSRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLITSGPVGS
Sbjct: 301 KSHPSSSSSQLSTGPARSRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLITSGPVGS 480
Query: 121 GPVGASRRSGQLEQSGSMGKAVYGSAVTSLGEEVKVGFRVSRS 163
GPVGASRRSGQLEQSGSMGKAVYGSAVTSLGEEVKVGFRVSRS
Sbjct: 481 GPVGASRRSGQLEQSGSMGKAVYGSAVTSLGEEVKVGFRVSRS 609
>TC78774 similar to GP|15292907|gb|AAK92824.1 unknown protein {Arabidopsis
thaliana}, partial (61%)
Length = 1375
Score = 271 bits (692), Expect(2) = 1e-76
Identities = 174/394 (44%), Positives = 220/394 (55%), Gaps = 53/394 (13%)
Frame = +1
Query: 1 MGTRIPSHQLSSGLYVSGRPEQPKERQPPTMASRSVPYTGGDPKKSGELGKMLDIPVLDP 60
MG+RIPSHQLS+GLYVSGRPEQPKER P TM + ++PYTGGD KKSGELGKM DIP
Sbjct: 241 MGSRIPSHQLSNGLYVSGRPEQPKERVP-TMTATAMPYTGGDIKKSGELGKMFDIPTDGS 417
Query: 61 KSHPS-----SSSSQLSTGPARSR-----PNS---------------GQVGKNI---TGS 92
KS S + S S G A S PNS G G N + S
Sbjct: 418 KSRKSGPITGAPSRTTSFGGAGSHSGPIHPNSAARAIYTTSGSMSIGGAAGSNSQKKSNS 597
Query: 93 GTLSR------KSTG-------------SGPI--ALQPTGLITSGPVGSGPV---GASRR 128
G LS+ KS+G SGP+ L TGLITSGP+ SGP+ GA R+
Sbjct: 598 GPLSKHGEPIKKSSGPQSGGVTPTGRQHSGPLPPVLPTTGLITSGPISSGPLNSSGAPRK 777
Query: 129 -SGQLEQSGSMGKAVYGSAVTSLGEEVKVGFRVSRSVVWVFMVVVAMCLLVGVFLMVAVK 187
SG LE +GSM + GSAV + V V +V+ S
Sbjct: 778 VSGPLEATGSM--KLQGSAVPVHNQAVTVLNQVAASFT---------------------- 885
Query: 188 KNVILFALGGVIVPVLVLIIWNCVLGRKGLLGFVKRYPDAELRGAIDGQYVKVTGVVTCG 247
WN GR+ ++GF+ YPD+ELR A +GQ+VKV+GVVTCG
Sbjct: 886 --------------------WNTYWGRRAIMGFIANYPDSELRTAKNGQFVKVSGVVTCG 1005
Query: 248 SIPLESSYQRIPRCVYVSSELYEYKGWGGKSAHPKHRCFTWGSRYSEKYIADFYISDFQT 307
++PLESS+Q++PRCVY S+ LYEY+GW K+A+P HR +WG R E+ + DFYISDFQ+
Sbjct: 1006NVPLESSFQKVPRCVYTSTSLYEYRGWDSKAANPTHRRLSWGLRLLERRVVDFYISDFQS 1185
Query: 308 GLRALVKAGYGNKVAPFVKPTTVVDVTKENRELS 341
GLRALVK G+G +V P+V + +++V ELS
Sbjct: 1186GLRALVKTGHGARVTPYVDDSVLINVNPTKEELS 1287
Score = 33.9 bits (76), Expect(2) = 1e-76
Identities = 16/27 (59%), Positives = 21/27 (77%)
Frame = +3
Query: 343 NFLGWLADRKLSTDDRIMRLKEGHIKE 369
+F GW R LS+DDRI RL+EG++KE
Sbjct: 1296 SFGGW-EKRNLSSDDRITRLEEGYVKE 1373
>TC83359 weakly similar to PIR|G86385|G86385 hypothetical protein AAG12737.1
[imported] - Arabidopsis thaliana, partial (7%)
Length = 783
Score = 38.5 bits (88), Expect = 0.005
Identities = 43/163 (26%), Positives = 62/163 (37%), Gaps = 41/163 (25%)
Frame = +3
Query: 28 PPTMASRSVPYTGG-----DPKKSGELGKMLDIPVLDPKSHPSSSSSQLSTGPARS---- 78
P T+ VP T G +P G L +P L S PSS S + T +
Sbjct: 279 PATVKVEQVPVTSGPAFSHNPSVPRATGTGLGVPSLQTSS-PSSVSQDIMTSNENAMDTK 455
Query: 79 ---------RP-NSGQVGKNI-------------TGSGTLSRKSTGSGPIALQPTGLITS 115
RP N Q NI +G ++ +S G P+A+ + +I+S
Sbjct: 456 PIVSMLQPIRPVNPAQANVNILNNLSQARQVMALSGGTSMGLQSMGQTPVAMHMSNMISS 635
Query: 116 GPVGSGPVGAS---------RRSGQLEQSGSMGKAVYGSAVTS 149
G SGP G + SG L S +G+ S++TS
Sbjct: 636 GTTSSGPTGQNVFSSGPSVITSSGSLTASAQVGQNSGLSSLTS 764
>TC77949 weakly similar to GP|11762218|gb|AAG40387.1 AT4g27520 {Arabidopsis
thaliana}, partial (34%)
Length = 1297
Score = 38.1 bits (87), Expect = 0.007
Identities = 46/147 (31%), Positives = 54/147 (36%), Gaps = 15/147 (10%)
Frame = +3
Query: 20 PEQPKERQPPTMASRSVPY---TGGDPKKSGELGKMLDIPVLD---PKSHPSSSSSQ--L 71
P PK P+ S P T P SG + IP P S P++SS +
Sbjct: 489 PRTPKSSPSPSAGGLSPPSPSPTTTTPSPSGSPPSPVAIPPASSPVPTSGPTASSPSPVV 668
Query: 72 ST----GPARSRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTG---LITSGPVGSGPVG 124
ST GP S P S V G S S GP AL P G GP GP G
Sbjct: 669 STPPAGGPMASSP-SPVVSTPPAGGPMASSPSPAGGPPALSPAGGPSTAAGGPPAPGPGG 845
Query: 125 ASRRSGQLEQSGSMGKAVYGSAVTSLG 151
A+ G G G + G A + G
Sbjct: 846 AATSPG----PGGAGASGPGGAAAAPG 914
>TC78878 similar to GP|4519792|dbj|BAA75744.1 Asp1 {Arabidopsis thaliana},
partial (53%)
Length = 1839
Score = 33.1 bits (74), Expect = 0.21
Identities = 18/54 (33%), Positives = 32/54 (58%), Gaps = 2/54 (3%)
Frame = +3
Query: 163 SVVWVFMVVVAMCLLVGVFLMVAVK--KNVILFALGGVIVPVLVLIIWNCVLGR 214
S +W+ +V + +++G+ +MV V V++ LGG+ VPV+ L + VL R
Sbjct: 504 SHLWLLRRMVVVGMIIGIMMMVIVMVMDRVVVVILGGIRVPVM*LDLLEMVLHR 665
>TC87810 similar to GP|21592752|gb|AAM64701.1 unknown {Arabidopsis
thaliana}, partial (52%)
Length = 1094
Score = 32.7 bits (73), Expect = 0.28
Identities = 30/108 (27%), Positives = 50/108 (45%), Gaps = 3/108 (2%)
Frame = +3
Query: 56 PVLDPKSHPSSSSSQLSTGPARSRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLIT- 114
P + ++PSS+S +G + + P++G G TG GT STG+G TG+ T
Sbjct: 453 PSVSGCAYPSSASG---SGTSTTSPSTG-TGTG-TGMGTTPSTSTGTGTGTSTGTGMGTG 617
Query: 115 --SGPVGSGPVGASRRSGQLEQSGSMGKAVYGSAVTSLGEEVKVGFRV 160
+G G+ P + G L G +G + S + ++ G R+
Sbjct: 618 TSTGATGNTPYSTTAPGGVL---GGIGSGMGPSGAGGMNDDSHGGLRL 752
>TC80856 similar to GP|15706274|emb|CAC69997. HMG I/Y like protein {Glycine
max}, partial (35%)
Length = 1552
Score = 32.3 bits (72), Expect = 0.36
Identities = 26/82 (31%), Positives = 35/82 (41%)
Frame = +2
Query: 50 GKMLDIPVLDPKSHPSSSSSQLSTGPARSRPNSGQVGKNITGSGTLSRKSTGSGPIALQP 109
G L+ LDP S LST P+R R GS S++ G P +QP
Sbjct: 662 GDELEFSRLDPNSRVVRVPVSLSTTPSRGRGRP-------PGSKAKSKRRPGRPP-KIQP 817
Query: 110 TGLITSGPVGSGPVGASRRSGQ 131
L++ VG G+ RR G+
Sbjct: 818 ETLVSGATVGVVLGGSKRRPGR 883
>TC88788 weakly similar to SP|P05790|FBOH_BOMMO Fibroin heavy chain
precursor (Fib-H) (H-fibroin). [Silk moth] {Bombyx
mori}, partial (2%)
Length = 782
Score = 32.0 bits (71), Expect = 0.47
Identities = 27/88 (30%), Positives = 35/88 (39%)
Frame = +3
Query: 66 SSSSQLSTGPARSRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLITSGPVGSGPVGA 125
S S ST + S S G GS SR +G+G A G GSG GA
Sbjct: 252 SGSGSASTSGSGSGSGSSGAGSE-AGSHAGSRAGSGAGSGAGSEAGSYAGSHAGSGSSGA 428
Query: 126 SRRSGQLEQSGSMGKAVYGSAVTSLGEE 153
+G ++GS + GS + G E
Sbjct: 429 GSEAG--SEAGSYAGSHAGSGSSRAGSE 506
>TC86615 weakly similar to GP|8778409|gb|AAF79417.1| F16A14.5 {Arabidopsis
thaliana}, partial (31%)
Length = 1278
Score = 30.8 bits (68), Expect = 1.0
Identities = 28/100 (28%), Positives = 40/100 (40%), Gaps = 6/100 (6%)
Frame = +2
Query: 28 PPTMASRSVPYTGGDPKKSGELGKMLDIPVLDPKSHPSSSSSQLSTGPARSRPNSGQ--- 84
PPT ++ S Y P G P + S S+ T P + P++G
Sbjct: 485 PPTSSTSSCAY----PSSGNTGGGTTTTPATNTPVGISPPSTVTPTTPTGTTPSTGTGTP 652
Query: 85 --VGKNI-TGSGTLSRKSTGSGPIALQPTGLITSGPVGSG 121
G TG+GT + TG+G PTG T +G+G
Sbjct: 653 TGTGTGTGTGTGTGTPTGTGTGTGMGTPTGTGTGTGIGTG 772
>TC88496 similar to GP|1079652|gb|AAD09209.1| late embryogenesis abundant
protein {Glycine soja}, partial (96%)
Length = 874
Score = 30.4 bits (67), Expect = 1.4
Identities = 29/125 (23%), Positives = 51/125 (40%), Gaps = 22/125 (17%)
Frame = +1
Query: 25 ERQPPTMASRSVPYTGGDP--------KKSGELGKMLDIPVLDPKSHPSSSSSQLSTGPA 76
E+ T+ ++ T DP KK ++ + ++ L + H +++ + +T A
Sbjct: 169 EKTKATLQEKTEKMTARDPVQKEMATHKKEAKMNQA-ELDKLAAREH--NAAVKQTTTAA 339
Query: 77 RSRPNSGQVGKNITGSGTLSRKSTGS--------------GPIALQPTGLITSGPVGSGP 122
TG+GT + +TG+ G QPTG + G VGS P
Sbjct: 340 AGHMGQPHHTTGTTGTGTATYSTTGNYGHPTGAHQMSAMPGHGTGQPTGHVVDGVVGSHP 519
Query: 123 VGASR 127
+G +R
Sbjct: 520 IGTNR 534
>BG454018 similar to GP|1079652|gb| late embryogenesis abundant protein
{Glycine soja}, partial (75%)
Length = 586
Score = 30.0 bits (66), Expect = 1.8
Identities = 18/52 (34%), Positives = 25/52 (47%), Gaps = 14/52 (26%)
Frame = +3
Query: 90 TGSGTLSRKSTGS--------------GPIALQPTGLITSGPVGSGPVGASR 127
TG+GT + +TG+ G QPTG + G VGS P+G +R
Sbjct: 288 TGTGTATYSTTGNYGHPTGAHQMSAMPGHGTGQPTGHVVDGVVGSHPIGTNR 443
>TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclear
ribonucleoprotein [imported] - Arabidopsis thaliana,
partial (60%)
Length = 1836
Score = 30.0 bits (66), Expect = 1.8
Identities = 20/53 (37%), Positives = 26/53 (48%), Gaps = 7/53 (13%)
Frame = -2
Query: 95 LSRKSTGSGPIALQPT-------GLITSGPVGSGPVGASRRSGQLEQSGSMGK 140
+S S+ S +L PT GLITSG +GSG G +R G + GK
Sbjct: 413 VSAPSSSSESESLDPTSPFCGIGGLITSGRIGSGARGRTREEGAVGIGARDGK 255
>TC83133 similar to PIR|T06033|T06033 hypothetical protein T28I19.140 -
Arabidopsis thaliana, partial (16%)
Length = 906
Score = 29.6 bits (65), Expect = 2.3
Identities = 18/60 (30%), Positives = 30/60 (50%), Gaps = 3/60 (5%)
Frame = +3
Query: 53 LDIPVLDPKSHPSS---SSSQLSTGPARSRPNSGQVGKNITGSGTLSRKSTGSGPIALQP 109
L +P +P S +S S Q + P +S+P+ GK + GSG + G+ +A +P
Sbjct: 663 LPVPSKEPPSRNTSVRSSVGQAAQVPGKSKPSLSNGGKLVRGSGENRKPVAGARHLAPKP 842
>BQ751207 weakly similar to GP|604427|gb|AA ACOB protein {Emericella
nidulans}, partial (12%)
Length = 758
Score = 29.6 bits (65), Expect = 2.3
Identities = 21/62 (33%), Positives = 29/62 (45%), Gaps = 2/62 (3%)
Frame = +1
Query: 22 QPKERQPPTMASRSVPYTGGDPKKSGELGKMLDIPVLDPKSHPSSS--SSQLSTGPARSR 79
Q +R+P T+ S S P P + P P HP+ + SSQ S+ P RSR
Sbjct: 34 QWSKRKPSTLLSHSWP*ANPQPLPAQP-------PTSSPARHPTQTPMSSQNSSKPLRSR 192
Query: 80 PN 81
P+
Sbjct: 193PS 198
>TC88164 GP|3763933|gb|AAC64313.1| unknown protein {Arabidopsis thaliana},
partial (1%)
Length = 879
Score = 29.3 bits (64), Expect = 3.0
Identities = 20/52 (38%), Positives = 30/52 (57%)
Frame = -3
Query: 355 TDDRIMRLKEGHIKEGSTVSVMGVVRRHENVLMIVPPTEPVSTGCQWMRCLL 406
T+ RI+ KEG I+ TVS +G E V+M+V +E +S + +R LL
Sbjct: 283 TN*RILE*KEGKIERSMTVSSLG----SEKVVMVVVGSESLSLLLRLLRRLL 140
>BF003766 similar to PIR|S56032|S560 probable membrane protein YJL078c -
yeast (Saccharomyces cerevisiae), partial (2%)
Length = 576
Score = 29.3 bits (64), Expect = 3.0
Identities = 22/74 (29%), Positives = 33/74 (43%)
Frame = +2
Query: 32 ASRSVPYTGGDPKKSGELGKMLDIPVLDPKSHPSSSSSQLSTGPARSRPNSGQVGKNITG 91
+S S+ +GG S G + P L S+ SSSS + S + + G +
Sbjct: 311 SSSSLSASGGINSTSASTGISMITPALAASSNNCSSSSTPVVATSSSTTSLFKFGSSPVT 490
Query: 92 SGTLSRKSTGSGPI 105
S LS S+GS P+
Sbjct: 491 SAGLSVSSSGSKPL 532
>BG586302
Length = 607
Score = 28.9 bits (63), Expect = 4.0
Identities = 11/45 (24%), Positives = 26/45 (57%)
Frame = -2
Query: 154 VKVGFRVSRSVVWVFMVVVAMCLLVGVFLMVAVKKNVILFALGGV 198
+K+ FR+ R + W + + + + + + L+ +V KN++ F L +
Sbjct: 543 MKMKFRIFREICWYILTYIILFIKLTIKLINSV*KNILAFFLNKI 409
>TC80622 homologue to PIR|T05792|T05792 hypothetical protein M7J2.80 -
Arabidopsis thaliana, partial (82%)
Length = 1124
Score = 28.9 bits (63), Expect = 4.0
Identities = 13/43 (30%), Positives = 25/43 (57%), Gaps = 4/43 (9%)
Frame = +1
Query: 350 DRKLSTDDRIMRLKEGHIKEGSTVSVMGVV----RRHENVLMI 388
++ S DR+ R+K ++KEG SV G++ H ++L++
Sbjct: 205 EKDTSVADRLARMKVNYMKEGMRTSVEGILLVQEHNHPHILLL 333
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.136 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,132,377
Number of Sequences: 36976
Number of extensions: 183889
Number of successful extensions: 1180
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 1152
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1167
length of query: 428
length of database: 9,014,727
effective HSP length: 99
effective length of query: 329
effective length of database: 5,354,103
effective search space: 1761499887
effective search space used: 1761499887
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC122163.2


