

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122162.9 - phase: 0
(380 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
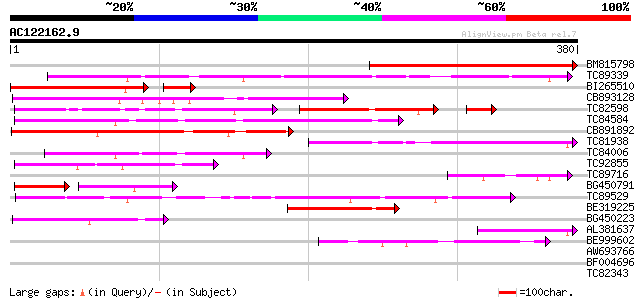
Score E
Sequences producing significant alignments: (bits) Value
BM815798 homologue to GP|7292702|gb|A CG15741 gene product {Dros... 281 2e-76
TC89339 similar to GP|17380986|gb|AAL36305.1 unknown protein {Ar... 257 5e-69
BI265510 weakly similar to PIR|C84555|C845 hypothetical protein ... 173 3e-50
CB893128 weakly similar to PIR|F96671|F966 hypothetical protein ... 187 8e-48
TC82598 weakly similar to PIR|C84547|C84547 hypothetical protein... 110 7e-44
TC84584 similar to PIR|C84547|C84547 hypothetical protein At2g17... 154 4e-38
CB891892 153 1e-37
TC81938 similar to PIR|C84547|C84547 hypothetical protein At2g17... 125 4e-29
TC84006 homologue to GP|9758228|dbj|BAB08727.1 gb|AAD22308.1~gen... 122 2e-28
TC92855 89 3e-18
TC89716 73 2e-13
BG450791 weakly similar to GP|9758228|dbj| gb|AAD22308.1~gene_id... 52 7e-12
TC89529 homologue to PIR|H86400|H86400 protein T17H3.7 [imported... 62 5e-10
BE319225 57 1e-08
BG450223 homologue to PIR|H71159|H711 hypothetical protein PH047... 57 2e-08
AL381637 weakly similar to PIR|C96738|C967 unknown protein F3I17... 54 1e-07
BE999602 weakly similar to GP|6686405|gb|A F1E22.11 {Arabidopsis... 53 2e-07
AW693766 similar to GP|16611746|gb| putative glycosyl transferas... 37 0.010
BF004696 37 0.016
TC82343 similar to GP|10177628|dbj|BAB10775. gb|AAF30317.1~gene_... 34 0.11
>BM815798 homologue to GP|7292702|gb|A CG15741 gene product {Drosophila
melanogaster}, partial (14%)
Length = 577
Score = 281 bits (720), Expect = 2e-76
Identities = 137/139 (98%), Positives = 137/139 (98%)
Frame = +3
Query: 242 PLGHDSPGGNKMLVESEGRLLLLGIEESSNYFSIDFFELDEKKKKWVRLMDFDEKEKKWV 301
PLG DSPG NKMLVESEGRLLLLGIEESSNYFSIDFFELDEKKKKWVRLMDFDEKEKKWV
Sbjct: 6 PLGLDSPGWNKMLVESEGRLLLLGIEESSNYFSIDFFELDEKKKKWVRLMDFDEKEKKWV 185
Query: 302 KLRNFGDRVFFIGRGCSFSASASDLCIQKGNCAIFIDESVLHNNNMVRGKRVFHLDQDRL 361
KLRNFGDRVFFIGRGCSFSASASDLCIQKGNCAIFIDESVLHNNNMVRGKRVFHLDQDRL
Sbjct: 186 KLRNFGDRVFFIGRGCSFSASASDLCIQKGNCAIFIDESVLHNNNMVRGKRVFHLDQDRL 365
Query: 362 SRGSKYLNLFLPPEWILKI 380
SRGSKYLNLFLPPEWILKI
Sbjct: 366 SRGSKYLNLFLPPEWILKI 422
>TC89339 similar to GP|17380986|gb|AAL36305.1 unknown protein {Arabidopsis
thaliana}, partial (3%)
Length = 1290
Score = 257 bits (657), Expect = 5e-69
Identities = 158/368 (42%), Positives = 217/368 (58%), Gaps = 16/368 (4%)
Frame = +3
Query: 26 DLIRFRSICSNWRSSSIPNHHNILPFKFPLLKCEDNSIKYNDNEISSSSNHH-------P 78
DLIRFRS+CS WRSSS+PNHH ILP+KFP+ K S + + N + P
Sbjct: 3 DLIRFRSVCSTWRSSSVPNHHYILPYKFPVFKLPFISEQNDKNPPFCHLSKQNIFLIKPP 182
Query: 79 SQLEEKQTLIPRRPWLIRFTQNSHGKTKLFHPLI-KYPRPQQEETLAHPRVFDFNKLSVL 137
Q +E+QTL+ RPWLIR +QN+ GKT+ FHPL P P+ + DF KLS+L
Sbjct: 183 QQQQEQQTLL--RPWLIRVSQNTRGKTRFFHPLFCDSPLPRIN------LILDFRKLSLL 338
Query: 138 HLGTDFILDKVNFTSRNR--HGIYMLPKTVLAVTCHGKDPLILGSLGYCSYRPLLFRDRD 195
HLG++ + S+++ + YM P+ V+AVTCHGK+PLI+ ++ PLL + +
Sbjct: 339 HLGSNTFTRDFDTYSQHQPFNSDYMFPEKVIAVTCHGKNPLIVATIS-SHPEPLLSKCGN 515
Query: 196 EHWKPIPKMSSLYGDTCVFKGQLYAVHQSGQTVTIGPDSSVELAAQPLGHDSPGGNKMLV 255
E WK IP MS +GD C+FKG+ YAV + G+T+ +G DS+V L A+PL D K+LV
Sbjct: 516 EKWKVIPDMSMEFGDICLFKGRAYAVDKIGKTIMVGSDSNVHLVAEPL--DGGENKKLLV 689
Query: 256 ESEGRLLLLGIEESSNYFSIDFFELDEKKKKWVRLMDFDEKEKKWVKLRNFGDRVFFIGR 315
ES+G LLL + + S+D F+L+E KEKKWVKL + GD+V F+G
Sbjct: 690 ESDGDLLLACV-HAFPVISVDLFKLNE-------------KEKKWVKLTSLGDKVLFLGI 827
Query: 316 GCSFSASASDLCIQKGNCAIFIDESVLHNNNMVRGKRVFHLDQDR-----LSRGSKYLNL 370
CSFSAS SDL + KGNC I I + + + + + LD D+ L +Y NL
Sbjct: 828 KCSFSASFSDLYVAKGNCVI-ISADIFRRLSRLGSQASYVLDLDQGWLPLLCDSPEYSNL 1004
Query: 371 FL-PPEWI 377
F PP+WI
Sbjct: 1005FWPPPKWI 1028
>BI265510 weakly similar to PIR|C84555|C845 hypothetical protein At2g17690
[imported] - Arabidopsis thaliana, partial (8%)
Length = 545
Score = 173 bits (438), Expect(2) = 3e-50
Identities = 88/111 (79%), Positives = 89/111 (79%), Gaps = 18/111 (16%)
Frame = +2
Query: 1 MAEADWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPNHHNILPFKFPLLKCED 60
MAEADWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPNHHNILPFKFPLLKCED
Sbjct: 116 MAEADWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPNHHNILPFKFPLLKCED 295
Query: 61 NSIKYNDNEISSSSNH------------------HPSQLEEKQTLIPRRPW 93
NSIKYNDNEI S N+ PSQLEEKQTLIPRRPW
Sbjct: 296 NSIKYNDNEIIDSINNTDSLFCYLSKRSIFLVKPPPSQLEEKQTLIPRRPW 448
Score = 43.5 bits (101), Expect(2) = 3e-50
Identities = 19/21 (90%), Positives = 19/21 (90%)
Frame = +1
Query: 104 KTKLFHPLIKYPRPQQEETLA 124
K LFHPLIKYPRPQQEETLA
Sbjct: 481 KPNLFHPLIKYPRPQQEETLA 543
Score = 40.4 bits (93), Expect = 0.001
Identities = 22/43 (51%), Positives = 25/43 (57%)
Frame = +3
Query: 68 NEISSSSNHHPSQLEEKQTLIPRRPWLIRFTQNSHGKTKLFHP 110
N +SSSSNHH + + LIRFTQNSHGKTK P
Sbjct: 372 NAVSSSSNHHHHN*RKNKP*FLVVLGLIRFTQNSHGKTKPLSP 500
>CB893128 weakly similar to PIR|F96671|F966 hypothetical protein F13O11.14
[imported] - Arabidopsis thaliana, partial (8%)
Length = 768
Score = 187 bits (474), Expect = 8e-48
Identities = 112/246 (45%), Positives = 147/246 (59%), Gaps = 21/246 (8%)
Frame = +3
Query: 3 EADWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPNHH-NILPFKFPLLKCEDN 61
E DWS+LPK+LL LISQ ID E+DLI FRSICS WRSSSIPNHH NI FKFP
Sbjct: 60 EVDWSELPKELLNLISQCIDNELDLIHFRSICSTWRSSSIPNHHSNISSFKFPKYIPPPY 239
Query: 62 SIKYNDNEISS--SSNHHPSQLEEKQTL----------IPRRPWLIRFTQ----NSHGKT 105
S K +DN+I +++ P K++L + RPWL++ N GK
Sbjct: 240 STKNDDNDIIHFIKNSNSPFDSFSKRSLFLVKAPPHQTVVLRPWLMQICPSSLPNKRGKI 419
Query: 106 KLF--HPLIKYPRPQQ--EETLAHPRVFDFNKLSVLHLGTDFILDKVNFTSRNRHGIYML 161
L HP I++ +Q E + V DFNKLSVLHLGT +I N++G
Sbjct: 420 MLTYSHPPIRHESAKQLQETLIERADVLDFNKLSVLHLGTRYI--------SNQYG--KC 569
Query: 162 PKTVLAVTCHGKDPLILGSLGYCSYRPLLFRDRDEHWKPIPKMSSLYGDTCVFKGQLYAV 221
P+T LAVTCHGK+PL+LG+L + S+ P++FR +E WK +S++ GD C+FKG+ Y V
Sbjct: 570 PETALAVTCHGKNPLLLGTLSHFSHLPVIFRGCNEQWKLFSNLSTVRGDICLFKGRFYVV 749
Query: 222 HQSGQT 227
SG+T
Sbjct: 750 EMSGRT 767
>TC82598 weakly similar to PIR|C84547|C84547 hypothetical protein At2g17030
[imported] - Arabidopsis thaliana, partial (2%)
Length = 1050
Score = 110 bits (274), Expect(3) = 7e-44
Identities = 74/178 (41%), Positives = 92/178 (51%), Gaps = 2/178 (1%)
Frame = +1
Query: 4 ADWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPNHHNILPFKFPLLKCEDNSI 63
ADWS L KDLL LIS R+D E D+IRFRS+CS WR IPNH LPFK P S
Sbjct: 70 ADWSTLHKDLLLLISSRLDNEADIIRFRSVCSTWR-FFIPNHS--LPFKLPHFHTSSLSC 240
Query: 64 KYNDNEISSSSNHHPSQLEEKQTLIPRRPWLIRFTQNSHGKTKLFHPLIKYPRPQQEETL 123
+ + I P Q E++QTL RPWLIR +NS +T+LFHPL R
Sbjct: 241 YLSKHNIFLIK---PPQQEQQQTL---RPWLIRIAKNSCRETQLFHPL----RSVDSSPY 390
Query: 124 AHPRVFDFNKLSVLHLGTDFILDKVN--FTSRNRHGIYMLPKTVLAVTCHGKDPLILG 179
P V D K SV+ LG D ++ + N T R + + + H +DP LG
Sbjct: 391 HFPHVLDLKKFSVVRLGADVVMGRYNEDKTVLERSKELLSQFELYQLKIHRQDPTPLG 564
Score = 72.8 bits (177), Expect(3) = 7e-44
Identities = 42/100 (42%), Positives = 60/100 (60%), Gaps = 7/100 (7%)
Frame = +2
Query: 195 DEHWKPIPKMSSLYGDTCVFKGQLYAVHQSGQTVTIGPDSSVELAAQPLGHDSPGGNKML 254
++HW IP MS+ + D CVFK + YAV++ G+TV G D SVEL A+ + + G K L
Sbjct: 677 EKHWTLIPCMSTYFLDICVFKERFYAVNKIGRTVAFGLDYSVELVAE---YVNGGDMKFL 847
Query: 255 VESEGRLLLLGIEESSNY-------FSIDFFELDEKKKKW 287
VE EG LLL+ I +S + + F L+E+KK+W
Sbjct: 848 VECEGELLLVDICDSHCFGFPGEKGLKLKVFRLNERKKEW 967
Score = 33.1 bits (74), Expect(3) = 7e-44
Identities = 15/20 (75%), Positives = 16/20 (80%)
Frame = +3
Query: 307 GDRVFFIGRGCSFSASASDL 326
G V F+G GCSFSASASDL
Sbjct: 984 GIGVLFLGNGCSFSASASDL 1043
>TC84584 similar to PIR|C84547|C84547 hypothetical protein At2g17030
[imported] - Arabidopsis thaliana, partial (4%)
Length = 812
Score = 154 bits (390), Expect = 4e-38
Identities = 101/274 (36%), Positives = 151/274 (54%), Gaps = 13/274 (4%)
Frame = +3
Query: 4 ADWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPNHHNILPFKFPLLKCEDNSI 63
ADWSQLPKDLL LIS ++D+E +RFRS+CS+WRSS NHH+ L L D++
Sbjct: 33 ADWSQLPKDLLQLISSKLDSEFYQLRFRSVCSSWRSSVPKNHHHHLTLPSHLPTPSDSNN 212
Query: 64 KYNDNE------------ISSSSNHHPSQLEEKQTLIPRRPWLIRFTQNSHGKTKLFHPL 111
++ I+ +NHH Q Q + PWLI+ +S +T+L++PL
Sbjct: 213 LHHSKYITFPLSKRTIFLITPPTNHHHHQ----QQTLNNNPWLIKIGPDSRDRTRLWNPL 380
Query: 112 IKYPRPQQEETLAHPRVFDFNKLSVLHLGTDFILDKVNFTSRNRHGIYMLPKTVL-AVTC 170
+ ++ ++ P+V +FN+ V+ LG +F++ N S +YM VL A
Sbjct: 381 SR----DKQLSIYLPQVINFNEHRVIDLGREFVIGNFNEYS----SLYMEKVIVLDADMW 536
Query: 171 HGKDPLILGSLGYCSYRPLLFRDRDEHWKPIPKMSSLYGDTCVFKGQLYAVHQSGQTVTI 230
GK+ + + S + LFR DE W +P+M S + D CVFKG+ AV +G+TV +
Sbjct: 537 GGKERCSVLFTIHISGKIALFRCGDERWTIVPEMPSPFDDVCVFKGRPVAVDGTGRTVAL 716
Query: 231 GPDSSVELAAQPLGHDSPGGNKMLVESEGRLLLL 264
PD S++L A+P+ G K LVES+G LLL+
Sbjct: 717 RPDLSLDLVAEPV---FGGDKKFLVESDGELLLV 809
>CB891892
Length = 587
Score = 153 bits (386), Expect = 1e-37
Identities = 94/200 (47%), Positives = 120/200 (60%), Gaps = 11/200 (5%)
Frame = +2
Query: 2 AEADWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPNHH-NILPFKFPLLK--- 57
A +DWS LP DL+ LISQ IDTEIDLIRFRS+C NWRSSSI HH NILPF+FPL K
Sbjct: 26 AMSDWSALPMDLVNLISQGIDTEIDLIRFRSVCCNWRSSSIRKHHLNILPFEFPLFKFRS 205
Query: 58 -----CEDNSIKYNDNEISSSSNHHPSQLEEKQTLIPRRPWLIRFTQNSHGKTKLFHPLI 112
DN+I + S E++QTLI RPWL+R Q S+GKT++ +PLI
Sbjct: 206 LFDSIINDNTIPFFCYLSKCSFFLIKPPHEQQQTLIRHRPWLVRIRQYSNGKTQILNPLI 385
Query: 113 KYPRPQQEETLAHPRVFDFNKLSVLHLGTDFIL--DKVNFTSRNRHGIYMLPKTVLAVTC 170
Y P DFNKLSV+HLGTDFI+ D + + YM P+ ++A T
Sbjct: 386 SYQSP-----FDSLHSIDFNKLSVIHLGTDFIVHDDSLLYD-------YMYPEKIVAFTG 529
Query: 171 HGKDPLILGSLGYCSYRPLL 190
+ K P++LG+ S +P+L
Sbjct: 530 NEKKPIVLGAFTTTS-KPIL 586
>TC81938 similar to PIR|C84547|C84547 hypothetical protein At2g17030
[imported] - Arabidopsis thaliana, partial (1%)
Length = 916
Score = 125 bits (313), Expect = 4e-29
Identities = 79/186 (42%), Positives = 106/186 (56%), Gaps = 6/186 (3%)
Frame = +1
Query: 201 IPKMSSLYG-DTCVFKGQLYAVHQSGQTVTIGPDSSVELAAQPLGHDSPGGNKMLVESEG 259
IP MS+ +G + CVFKG+ V ++G+T IGPD + L A+P+ G +K LVES
Sbjct: 1 IPNMSNSHGGNVCVFKGRPCVVDKTGRTFMIGPDLTTHLIAEPV---FGGKSKFLVESSE 171
Query: 260 RLLLLGIEESSNYFSIDFFELDEKKKKWVRLMDFDEKEKKWVKLRNFGDRVFFIGRGCSF 319
LLL ++ NY + W+ ++ DEKEK+WVKL N GD+V F+G G SF
Sbjct: 172 FELLL-VDRYENY----------RVPVWIDVLRLDEKEKRWVKLANLGDKVLFLGNGYSF 318
Query: 320 SASASDLCIQKGNCAIFIDESVLHNNNMVRGK-RVFHLDQDRLSRGSKYLNLFL----PP 374
SASAS+L GNC I+ H+ N+ + K VFHLDQ + S S Y F PP
Sbjct: 319 SASASELGFANGNCVIYSKSYGFHDLNIRKCKMSVFHLDQGQASPLSDYPEYFKLFWPPP 498
Query: 375 EWILKI 380
EWI K+
Sbjct: 499 EWIAKL 516
>TC84006 homologue to GP|9758228|dbj|BAB08727.1
gb|AAD22308.1~gene_id:MZF18.8~similar to unknown protein
{Arabidopsis thaliana}, partial (4%)
Length = 597
Score = 122 bits (306), Expect = 2e-28
Identities = 70/159 (44%), Positives = 94/159 (59%), Gaps = 7/159 (4%)
Frame = +2
Query: 24 EIDLIRFRSICSNWRSSSIPNHHNILPFKFPLLKCEDNSIKYNDNE----ISSSSNHHPS 79
E+DLI FRS+CS WRSSS+P HH F+FPLLK N N N+ I+ +
Sbjct: 137 EVDLIHFRSVCSTWRSSSVPKHHRNFRFQFPLLKFPFNPDFINRNKSFCYITKQNIFLIK 316
Query: 80 QLEEKQTLIPRRPWLIRFTQNSHGKTKLFHPLIKYPRPQQEETLAHPRVFDFNKLSVLHL 139
+++QTL RP LIR QN+ GK+KL+HPL+ Q + + DFNKLS+LHL
Sbjct: 317 PPQQEQTL-NLRPSLIRICQNARGKSKLYHPLL-----QNGDKFLYNFALDFNKLSILHL 478
Query: 140 GTDFILDKVNFTSRNR---HGIYMLPKTVLAVTCHGKDP 175
G++F + +FT ++ YM PK V+ VTCHGK P
Sbjct: 479 GSNFFMIDFDFTFNDKLYNPDYYMYPKKVVVVTCHGKKP 595
>TC92855
Length = 625
Score = 89.0 bits (219), Expect = 3e-18
Identities = 59/146 (40%), Positives = 73/146 (49%), Gaps = 9/146 (6%)
Frame = +2
Query: 4 ADWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPN--HHNILPFKFPLLKCEDN 61
ADWS LP +LL LISQ++DT + L+RFRS+C+ WRSSSIPN H N P P E N
Sbjct: 53 ADWSHLPSELLQLISQKLDTVLCLLRFRSVCATWRSSSIPNHKHKNSSPLNIPQFP-EPN 229
Query: 62 SIKYNDNEISSSS-------NHHPSQLEEKQTLIPRRPWLIRFTQNSHGKTKLFHPLIKY 114
+N I S H ++ Q PW IR + GKT+L+HP
Sbjct: 230 FDPFNIRNIIKSKYITSRLITHTIFLIKPPQHQPSLNPWWIRIGPDFKGKTQLWHPF--- 400
Query: 115 PRPQQEETLAHPRVFDFNKLSVLHLG 140
Q + V DFNKLS LG
Sbjct: 401 -SSGQNLSSDFNNVLDFNKLSFRVLG 475
>TC89716
Length = 735
Score = 72.8 bits (177), Expect = 2e-13
Identities = 48/99 (48%), Positives = 56/99 (56%), Gaps = 15/99 (15%)
Frame = +2
Query: 294 DEKEKKWVKLRNFGDRVFFIGRG--CSFSASASDLCIQKGNCAIFIDESVLHNNNMVRGK 351
+EKEKKWVKL + GDR+ F+G G CSFSA ASDLC+ KGN SV+ NN +
Sbjct: 8 NEKEKKWVKLTSLGDRILFLGLGLVCSFSACASDLCVAKGN-------SVIFNNYIFESY 166
Query: 352 R---------VFHLDQDR---LSRGSKYLNLFLPP-EWI 377
R V LDQ R LS +Y NL PP EWI
Sbjct: 167 RPFGFQCKQYVLDLDQGRHSLLSDCPEYSNLLWPPAEWI 283
>BG450791 weakly similar to GP|9758228|dbj|
gb|AAD22308.1~gene_id:MZF18.8~similar to unknown protein
{Arabidopsis thaliana}, partial (9%)
Length = 646
Score = 51.6 bits (122), Expect(2) = 7e-12
Identities = 23/37 (62%), Positives = 30/37 (80%)
Frame = -2
Query: 4 ADWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSS 40
A+WSQLP + L LISQ+++ E+ LIR RS+CS WRSS
Sbjct: 366 AEWSQLPHEFLQLISQKLNRELYLIRSRSVCSWWRSS 256
Score = 36.2 bits (82), Expect(2) = 7e-12
Identities = 22/68 (32%), Positives = 32/68 (46%), Gaps = 2/68 (2%)
Frame = -1
Query: 47 NILPFKFPLLKCEDNSIKYNDNEISSSSNHHPSQLE--EKQTLIPRRPWLIRFTQNSHGK 104
N +P KF C+ N + + H + + Q + +RPWLIR + GK
Sbjct: 235 NYIPVKFGQFSCQLFRSHPNIDVVMYLIKHDIFLFKPPKHQQNLHQRPWLIRIGPDFDGK 56
Query: 105 TKLFHPLI 112
TKL HPL+
Sbjct: 55 TKLLHPLL 32
>TC89529 homologue to PIR|H86400|H86400 protein T17H3.7 [imported] -
Arabidopsis thaliana, partial (1%)
Length = 1745
Score = 61.6 bits (148), Expect = 5e-10
Identities = 82/346 (23%), Positives = 142/346 (40%), Gaps = 11/346 (3%)
Frame = +1
Query: 5 DWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPNHHNILPFKFPLLKCEDNSIK 64
+W++LP++++ IS+R+ D IRFR +C W +SS+P LP + P+L D S
Sbjct: 451 EWAELPQEIIESISKRLTIYADYIRFRCVCRTW-NSSVPKTPLHLPPQLPILMLSDESF- 624
Query: 65 YNDNEISSSSNHH---PSQLEEKQTLIPRRPWLIRFTQNSHGKTKLFHPLIKYPRPQQEE 121
++S++ H P + WL+ + S+ + P
Sbjct: 625 ---FDLSTNKIHRLNFPLPSGPTRICGSSHGWLVILDETSN---------LNLLNPVTHA 768
Query: 122 TLAHPRVFDFNKLSVLHLGTDFILDKVNFTSRNRHGIYMLPKTVLAVTCHGKDPLILGSL 181
TL+ P + F +++ K+ + N++ +Y++ K VL+ D +
Sbjct: 769 TLSLPSLHTF----------PYLVKKL--FAFNKNNLYLI-KVVLSSNPSPNDD--FAAF 903
Query: 182 GYCSYRPLLF-RDRDEHWKPIPKMSSLYGDTCVFK-GQLYAVHQSGQT----VTIGPDSS 235
R L F R + W + + V+K G +A+ SG T V GP S
Sbjct: 904 AILGLRHLAFCRKGCDSWVLLDANVGDHWTDAVYKNGSFFAMSTSGITAVCDVVEGPRVS 1083
Query: 236 VELAAQPLGHDSPGGNKMLVESEGRLLLLGIEESSNYFSIDFFELDEKK--KKWVRLMDF 293
L ++ V S +LL+ + Y DF + K W+ M++
Sbjct: 1084YIPPPTSLYYNDV---FYAVFSGEEMLLIQRCCTEEYEPRDFEPYPDMSTVKFWIYKMNW 1254
Query: 294 DEKEKKWVKLRNFGDRVFFIGRGCSFSASASDLCIQKGNCAIFIDE 339
+ KW +++ G+ FIG+ S S SA+D NC F D+
Sbjct: 1255N--MLKWEEIQTLGEHSLFIGKNYSLSFSAADFAGCCPNCIYFTDD 1386
>BE319225
Length = 242
Score = 57.0 bits (136), Expect = 1e-08
Identities = 33/77 (42%), Positives = 51/77 (65%), Gaps = 2/77 (2%)
Frame = +2
Query: 187 RPLLFRDRDEHWKPIPKMSSLYGDTCVFKGQLYAVHQSGQTVTIG-PDSSVELAAQPLGH 245
+P+L + +E+ IP M++ + D C+FKG+ Y VH++G+T T+G D + EL A+ L
Sbjct: 17 KPILLKCSNENRNGIPVMTAWFEDICIFKGRPYVVHKTGRTATLGLDDFTFELVAESL-- 190
Query: 246 DSPGGN-KMLVESEGRL 261
S GG+ K LVES+G L
Sbjct: 191 VSGGGDIKFLVESDGDL 241
>BG450223 homologue to PIR|H71159|H711 hypothetical protein PH0477 -
Pyrococcus horikoshii, partial (8%)
Length = 604
Score = 56.6 bits (135), Expect = 2e-08
Identities = 36/111 (32%), Positives = 55/111 (49%), Gaps = 7/111 (6%)
Frame = +1
Query: 3 EADWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPNHHNILPFK-------FPL 55
+ DWS LPK+L + I + +D ID++RFRS+C ++RSS P+H N F F
Sbjct: 271 KVDWSHLPKELWSKIGKYLDNHIDILRFRSVCESFRSSIPPSHPNSPSFPLQIPHTIFDS 450
Query: 56 LKCEDNSIKYNDNEISSSSNHHPSQLEEKQTLIPRRPWLIRFTQNSHGKTK 106
L S Y + SSN + L T + WLI+ ++++ K
Sbjct: 451 LYYLHQSTIYLIEPTNGSSNFNLPPLAPSYT----KGWLIKXEESNNNPLK 591
>AL381637 weakly similar to PIR|C96738|C967 unknown protein F3I17.1
[imported] - Arabidopsis thaliana, partial (9%)
Length = 508
Score = 53.9 bits (128), Expect = 1e-07
Identities = 33/72 (45%), Positives = 40/72 (54%), Gaps = 5/72 (6%)
Frame = +3
Query: 314 GRGCSFSASASDLCIQKGNCAIFIDESVLHNNNMVRGK-RVFHLDQDRLSRGSKYLNLFL 372
G G SFSASAS+L GNC I+ H+ N+ + K VFHLDQ + S S Y F
Sbjct: 3 GNGYSFSASASELGFANGNCVIYSKSYGFHDLNIRKCKMSVFHLDQGQASPLSDYPEYFK 182
Query: 373 ----PPEWILKI 380
PPEWI K+
Sbjct: 183 LFWPPPEWIAKL 218
>BE999602 weakly similar to GP|6686405|gb|A F1E22.11 {Arabidopsis thaliana},
partial (4%)
Length = 579
Score = 53.1 bits (126), Expect = 2e-07
Identities = 45/176 (25%), Positives = 75/176 (42%), Gaps = 21/176 (11%)
Frame = +1
Query: 208 YGDTCVFKGQLYAVHQSGQTVTIGPDSSVELAAQPLGHDSP-----GGNKMLVESEGRLL 262
Y D VFKGQ Y + G S +++++ L SP G K LVES G L
Sbjct: 16 YDDLIVFKGQFYVTDKWGTI------SWIDISSLKLIQFSPPLCGFGNKKHLVESCGNLY 177
Query: 263 LL----------------GIEESSNYFSIDFFELDEKKKKWVRLMDFDEKEKKWVKLRNF 306
++ G + Y ++ F++ + ++W KWV +++
Sbjct: 178 VVDRYSYYEDENMGGNYDGRRHQARYEVVECFKVYKLDEEW----------GKWVDVKDL 327
Query: 307 GDRVFFIGRGCSFSASASDLCIQKGNCAIFIDESVLHNNNMVRGKRVFHLDQDRLS 362
+R F + C+FS SA +L +GNC F D + R+++LD R++
Sbjct: 328 KNRAFILSPSCNFSVSAKELIGYQGNCIYFKDSFSV---------RMYNLDDCRIT 468
>AW693766 similar to GP|16611746|gb| putative glycosyl transferase {Shigella
boydii}, partial (5%)
Length = 656
Score = 37.4 bits (85), Expect = 0.010
Identities = 20/49 (40%), Positives = 28/49 (56%)
Frame = +1
Query: 8 QLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPNHHNILPFKFPLL 56
QLP DLL +I + +D + DL +F S+C NWR N + + PLL
Sbjct: 346 QLPWDLLDIILRNLDFD-DLFQFGSVCKNWREFHKIYWRNFMASEEPLL 489
>BF004696
Length = 593
Score = 36.6 bits (83), Expect = 0.016
Identities = 39/144 (27%), Positives = 64/144 (44%), Gaps = 8/144 (5%)
Frame = +2
Query: 23 TEIDLIRFRSICSNWRS--SSIPNHHNI-LPFK-FPLLKCEDNSIKYNDNEISSSSNHHP 78
T ID++RFR+IC WRS + P HN+ +P + + LL+ + I+ S +H+P
Sbjct: 2 TTIDVVRFRTICRFWRSLVPAPPGSHNLCIPHQNYFLLQTKIYHIE------PSPHDHNP 163
Query: 79 SQLEEKQTLIPRRPWLIRFTQNSHGKTKLFHPLIKYPRPQQEETLAHPRVFDFNKLSVLH 138
S T + +I+ QNS+ L R + EE +V D V+
Sbjct: 164 S------TSCSNQGGIIKGFQNSNSSKLYLFDLFTNTRIRAEENT--EKVLDLMNFLVVE 319
Query: 139 LGTDFIL----DKVNFTSRNRHGI 158
L + DK+ F +R+ +
Sbjct: 320 LSEVYTRSHHEDKIGFKCASRNNV 391
>TC82343 similar to GP|10177628|dbj|BAB10775.
gb|AAF30317.1~gene_id:K6M13.17~similar to unknown
protein {Arabidopsis thaliana}, partial (9%)
Length = 666
Score = 33.9 bits (76), Expect = 0.11
Identities = 20/46 (43%), Positives = 27/46 (58%), Gaps = 1/46 (2%)
Frame = +2
Query: 1 MAEADWSQ-LPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPNH 45
MAE D LP +L+T I R+ + LIRF+S+C +W S NH
Sbjct: 38 MAEVDAPPYLPDELITKILVRLPVK-SLIRFKSVCKSWFSLISDNH 172
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.139 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,875,708
Number of Sequences: 36976
Number of extensions: 253845
Number of successful extensions: 1273
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 1233
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1240
length of query: 380
length of database: 9,014,727
effective HSP length: 98
effective length of query: 282
effective length of database: 5,391,079
effective search space: 1520284278
effective search space used: 1520284278
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC122162.9


