

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122162.14 - phase: 0
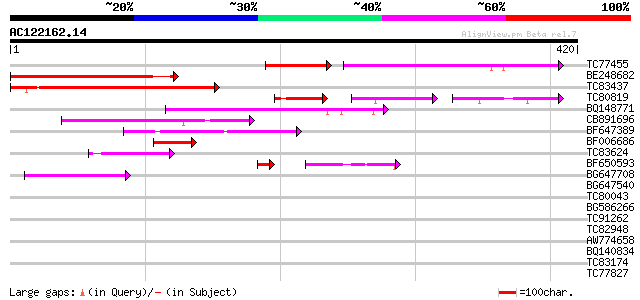
(420 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarot... 130 3e-47
BE248682 similar to GP|18568269|gb putative gag-pol polyprotein ... 157 6e-39
TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imp... 110 1e-24
TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non... 57 2e-20
BQ148771 71 7e-13
CB891696 67 1e-11
BF647389 52 3e-07
BF006686 48 8e-06
TC83624 homologue to PIR|G84581|G84581 copia-like retroelement p... 45 5e-05
BF650593 42 1e-04
BG647708 weakly similar to GP|13786450|gb| putative reverse tran... 44 2e-04
BG647540 40 0.001
TC80043 similar to GP|9759561|dbj|BAB11163.1 MtN21 nodulin prote... 37 0.014
BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-li... 37 0.014
TC91262 similar to GP|20804797|dbj|BAB92481. putative amino acid... 35 0.071
TC82948 34 0.092
AW774658 similar to GP|2808681|emb| Hcr9-4B {Lycopersicon hirsut... 28 1.0
BQ140834 30 2.3
TC83174 30 2.3
TC77827 homologue to GP|15131692|emb|CAC48393. putative aminoald... 29 3.0
>TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarotenoid
dioxygenase1 {Pisum sativum}, partial (43%)
Length = 1865
Score = 130 bits (326), Expect(2) = 3e-47
Identities = 73/175 (41%), Positives = 91/175 (51%), Gaps = 12/175 (6%)
Frame = -2
Query: 248 SWPSYISRWWKDVAFLEDEGGERWFNAEVVRKVGCGNSTSFWKDPWRGDIPFSRKYPRLF 307
+WP+Y SRWWKD+ LE+ G RWF E++RKVG G S+ FWKD W +P +PR F
Sbjct: 796 NWPAYASRWWKDLMSLEEVGRVRWFPRELIRKVGDGRSSFFWKDAWDSSVPLRESFPRAF 617
Query: 308 AISNNKETLVEECRQMNGVGGGWIFEWHR-PLFVWEEELLISLKEDLEG-------HRWV 359
+ + MN G W W R LF WE+E L+ L LEG WV
Sbjct: 616 FPYRLLKMGCGDLWDMNAEGVRWRLYWRRLELFEWEKERLLELLGRLEGVVLRYWADIWV 437
Query: 360 NDPDR----WVKSCYGKLERLLGGEVEWSLEELRVLESIWNSKAPLKVIAFSWKL 410
PD+ V SCY L+ L E S EE + +W SKAP KV+AFSW L
Sbjct: 436 WKPDKEGVFSVNSCYFLLQNLRLLEDRLSYEEEVIFRELWKSKAPAKVLAFSWTL 272
Score = 76.6 bits (187), Expect(2) = 3e-47
Identities = 35/49 (71%), Positives = 40/49 (81%)
Frame = -3
Query: 190 CQPRSKGGLGVRDIRVVNLSLLAKWRWRVLQGEEGLWKEVLIEKYGTRV 238
C PR KGGLGVRDIR+VN+SLLAKW WR+LQ + LWKEVL + YG RV
Sbjct: 969 CLPRCKGGLGVRDIRLVNVSLLAKWWWRLLQDQSSLWKEVLEDIYGPRV 823
>BE248682 similar to GP|18568269|gb putative gag-pol polyprotein {Zea mays},
partial (1%)
Length = 441
Score = 157 bits (398), Expect = 6e-39
Identities = 80/125 (64%), Positives = 92/125 (73%)
Frame = +3
Query: 1 MRNAVSRNLFKGFKIKNDGTGISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKV 60
M+NAV+RNLF+GF +K GT +SHLQYADDTLCIG +VDNLWTLKALL+GFEMASGLKV
Sbjct: 96 MKNAVNRNLFQGFDVKRGGTRVSHLQYADDTLCIGMPTVDNLWTLKALLQGFEMASGLKV 275
Query: 61 NFYKSCLMGINVPSEFMTMACDFLNCSEGVVPFKYLGLPVGANSFKLVTWEPLLEQLSRK 120
NF+KS L+GINVP +FM AC FLNC E +PF YLGLP G+ S K R
Sbjct: 276 NFHKSSLIGINVPRDFMEAACRFLNCREESIPFIYLGLPGGS*S*K------------RL 419
Query: 121 LHSWG 125
H WG
Sbjct: 420 SHLWG 434
>TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 951
Score = 110 bits (275), Expect = 1e-24
Identities = 59/157 (37%), Positives = 96/157 (60%), Gaps = 2/157 (1%)
Frame = +2
Query: 1 MRNAVSRNLFK--GFKIKNDGTGISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGL 58
M++ V LF F + N +SHLQ+A+DTL + + N+ L+A L F SGL
Sbjct: 476 MKSLVQTQLFTRYSFGVVNPVV-VSHLQFANDTLLLETKNWANIRALRAALVIF*AMSGL 652
Query: 59 KVNFYKSCLMGINVPSEFMTMACDFLNCSEGVVPFKYLGLPVGANSFKLVTWEPLLEQLS 118
KVNF+KS L+ +N+ +++ A L+ G VPF YLG+P+ NS +L WEP++ ++
Sbjct: 653 KVNFHKSGLVCVNIAPSWLSEAASVLSWKVGKVPFLYLGMPIEGNSRRLSFWEPIVNRIK 832
Query: 119 RKLHSWGNKYVSLGGRIVLLNAVINSIPIFYLSFLKM 155
+L W ++++S GGR+VLL +V+ S+ ++ L K+
Sbjct: 833 ARLTGWNSRFLSFGGRLVLLKSVLTSLSVYALPSSKL 943
>TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non-LTR
retroelement reverse transcriptase {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 1262
Score = 57.0 bits (136), Expect(3) = 2e-20
Identities = 26/67 (38%), Positives = 34/67 (49%), Gaps = 3/67 (4%)
Frame = +3
Query: 254 SRWWKDVAFLEDEGGE---RWFNAEVVRKVGCGNSTSFWKDPWRGDIPFSRKYPRLFAIS 310
S WWK + + + GE WF + VG G FW D W GD+P KYPRL ++
Sbjct: 177 SMWWKTICKVREGVGEGVGNWFEENIRMVVGDGRDAFFWYDTWAGDVPLRLKYPRLLDLA 356
Query: 311 NNKETLV 317
+KE V
Sbjct: 357 MDKECKV 377
Score = 43.5 bits (101), Expect(3) = 2e-20
Identities = 22/39 (56%), Positives = 25/39 (63%)
Frame = +1
Query: 197 GLGVRDIRVVNLSLLAKWRWRVLQGEEGLWKEVLIEKYG 235
GLGV NLSLL KW WR+L +EGLW VL +YG
Sbjct: 31 GLGVG---AFNLSLLGKWCWRLLVDKEGLWHRVLKARYG 138
Score = 36.2 bits (82), Expect(3) = 2e-20
Identities = 27/94 (28%), Positives = 42/94 (43%), Gaps = 12/94 (12%)
Frame = +2
Query: 329 GWIFEWHRPLFVWEEELL---ISLKEDLEGHRWVNDPDRWVKSCYGKLERLLGGEVE--- 382
G +EW R LFVWEEE + L + VND RW+ L+ + G V+
Sbjct: 413 GRAWEWTRRLFVWEEECVRECCILLNNFVLQDNVNDKWRWL------LDPVNGYSVKVFY 574
Query: 383 ------WSLEELRVLESIWNSKAPLKVIAFSWKL 410
+ + +++ +W+ P KV F W+L
Sbjct: 575 RYITSTGHISDRSLVDDVWHKHIPSKVSLFVWRL 676
>BQ148771
Length = 680
Score = 71.2 bits (173), Expect = 7e-13
Identities = 43/177 (24%), Positives = 84/177 (47%), Gaps = 12/177 (6%)
Frame = -3
Query: 116 QLSRKLHSWGNKYVSLGGRIVLLNAVINSIPIFYLSFLKMPNKVWRKIVKIQCDFLWGGA 175
Q+ L +W ++SL R+ L +VI ++P++ + +P +I K+Q F+WG
Sbjct: 588 QVHVMLANWKANHLSLARRVTLAKSVIEAVPLYPMMTTIIPKACIEEIQKLQRKFVWGDT 409
Query: 176 RGGKKLCWVKWRVVCQPRSKGGLGVRDIRVVNLSLLAKWRWRVLQGEEGLWKEVLIEKY- 234
++ V W + +P++ GLG+R + V+N + + K W + G L EV+ KY
Sbjct: 408 EVSRRYHAVGWETMSKPKTIYGLGLRRLDVMNKACIMKLGWSIYSGSNSLCTEVMRGKYQ 229
Query: 235 -GTRVCNLLVE---DDGSWPSYISRWWKDVAFLEDEGG-------ERWFNAEVVRKV 280
+ + +E D W + + W + L D G ++W +V++++
Sbjct: 228 RSESLEEIFLEKPTDSSLWKALVKLWPEIERNLVDSNGNWNWEKLKQWLPFDVLQRI 58
>CB891696
Length = 638
Score = 67.0 bits (162), Expect = 1e-11
Identities = 50/146 (34%), Positives = 76/146 (51%), Gaps = 3/146 (2%)
Frame = +1
Query: 39 VDNLWTLKALLRGFEMASGLKVNFYKSCLMGINVPSEFMTMACDFLNCSEGVVPFKYLGL 98
V+N+ T+K ++ FE+AS L VNF KS L+ +NV F ++ C V FKYLG+
Sbjct: 4 VENILTMKTIVSYFELASSLWVNFLKSGLINLNVIGHF*GW*NIYIKCKVH*VIFKYLGI 183
Query: 99 PVGANSFKLVTWEPLLEQLSRKLHSWGNK---YVSLGGRIVLLNAVINSIPIFYLSFLKM 155
VG N ++ E LL+ L+ L SW N + G + + +I Y S +K+
Sbjct: 184 LVGENPCRVNM*ELLLKLLTN*LGSWWNTK*LWTQNGFSQIHAK*ISQNI---YFSLMKI 354
Query: 156 PNKVWRKIVKIQCDFLWGGARGGKKL 181
P KV I +++ FL G + KK+
Sbjct: 355 PVKV*ELISQLKTQFL*GNLKVTKKI 432
>BF647389
Length = 404
Score = 52.4 bits (124), Expect = 3e-07
Identities = 32/133 (24%), Positives = 67/133 (50%), Gaps = 1/133 (0%)
Frame = +2
Query: 85 NCSEGVVPFKYLGLPVGANSFKLVTWEPLLEQLSRKLHSWGNKYVSLGGRIVLLNAVINS 144
N +G +P +YLG+P+ + ++ ++++ ++ + +K +S G + L+ V+
Sbjct: 14 NLIKGKLPSRYLGVPLSSKKLYVI---QRVKKIICRIEN*SSKLLSYAGSLQLIKIVLFG 184
Query: 145 IPIFYLSFLKMPNKVWRKIVKIQCD-FLWGGARGGKKLCWVKWRVVCQPRSKGGLGVRDI 203
+ ++ +P KV K+++ C FL G G K + +C P++ GG V D+
Sbjct: 185 VQPYWSQVFVLP*KV-IKLIQTTCRIFL*TGKSGTSKRALIAREHICLPKTAGGWNVIDL 361
Query: 204 RVVNLSLLAKWRW 216
+V N + + K W
Sbjct: 362 KVXNQTAICKLXW 400
>BF006686
Length = 325
Score = 47.8 bits (112), Expect = 8e-06
Identities = 21/32 (65%), Positives = 25/32 (77%)
Frame = +3
Query: 107 LVTWEPLLEQLSRKLHSWGNKYVSLGGRIVLL 138
L WEPLLE +++ L SWGNK +S GGRIVLL
Sbjct: 228 LPMWEPLLEHVNKMLKSWGNKLLSFGGRIVLL 323
>TC83624 homologue to PIR|G84581|G84581 copia-like retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(1%)
Length = 831
Score = 45.1 bits (105), Expect = 5e-05
Identities = 24/64 (37%), Positives = 38/64 (58%)
Frame = +1
Query: 59 KVNFYKSCLMGINVPSEFMTMACDFLNCSEGVVPFKYLGLPVGANSFKLVTWEPLLEQLS 118
+VNF M +N+ F+ + +FL C+ VPF +LGLP+GAN + T +P+L+ L
Sbjct: 262 RVNF-----MALNLEESFVEASPNFLLCNVNEVPFCFLGLPIGANPKRSSTRKPVLDSLQ 426
Query: 119 RKLH 122
L+
Sbjct: 427 WLLY 438
>BF650593
Length = 486
Score = 41.6 bits (96), Expect(2) = 1e-04
Identities = 25/77 (32%), Positives = 38/77 (48%), Gaps = 7/77 (9%)
Frame = +1
Query: 220 QGEEGLWKEVLIEKYGTRVCNLLVEDDGSWPSYISRWWKDVAFLEDEGGERWFNAEVVRK 279
+G GLW L+ KYG ++ +E+ G +S WWKD+ ++ D GG N V++
Sbjct: 73 RGVIGLWFRALVNKYGLNRGSITIENRG-----VSLWWKDICYI-DFGGVESLNVYSVKE 234
Query: 280 VGCG-------NSTSFW 289
V G +S S W
Sbjct: 235 VYKGLMSNVSSSSXSIW 285
Score = 21.6 bits (44), Expect(2) = 1e-04
Identities = 7/13 (53%), Positives = 9/13 (68%)
Frame = +2
Query: 184 VKWRVVCQPRSKG 196
VKW VC+P +G
Sbjct: 41 VKWEKVCRPLEEG 79
>BG647708 weakly similar to GP|13786450|gb| putative reverse transcriptase
{Oryza sativa}, partial (9%)
Length = 708
Score = 43.5 bits (101), Expect = 2e-04
Identities = 25/79 (31%), Positives = 39/79 (48%), Gaps = 1/79 (1%)
Frame = +1
Query: 12 GFKIKNDGTGISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKVNFYKS-CLMGI 70
G ++ I+HL +ADD+L A++ T+ +L ++ ASG VNF KS
Sbjct: 115 GIQVARSDPKITHLLFADDSLLFARANLTEAATIMQVLHSYQSASGQLVNFEKSEVSYSQ 294
Query: 71 NVPSEFMTMACDFLNCSEG 89
NVP++ M C + G
Sbjct: 295 NVPNQEKEMICQQIAIKTG 351
>BG647540
Length = 625
Score = 40.4 bits (93), Expect = 0.001
Identities = 30/99 (30%), Positives = 41/99 (41%), Gaps = 17/99 (17%)
Frame = +3
Query: 324 NGVGGGWIFEWHRPLFVWEEE--------LLISLKE---------DLEGHRWVNDPDRWV 366
N G W W R LF+WE + ++E DLE + W + V
Sbjct: 225 NSTDGYWDVNWRRSLFLWEFNTCFGSCWRIWTDIEEGKVRMYGGGDLEENGWFS-----V 389
Query: 367 KSCYGKLERLLGGEVEWSLEELRVLESIWNSKAPLKVIA 405
S Y KLE + E + ++V IW S AP KV+A
Sbjct: 390 NSMYKKLEGMRFEEGSLTEMRVKVFTHIWKSSAPSKVVA 506
>TC80043 similar to GP|9759561|dbj|BAB11163.1 MtN21 nodulin protein-like
{Arabidopsis thaliana}, partial (28%)
Length = 1421
Score = 37.0 bits (84), Expect = 0.014
Identities = 20/54 (37%), Positives = 28/54 (51%), Gaps = 1/54 (1%)
Frame = +1
Query: 269 ERWFNAEVVRKVGCGNSTSFWKDPW-RGDIPFSRKYPRLFAISNNKETLVEECR 321
+ W + + + V G TSFW + W RG R+Y RLF I +KE V + R
Sbjct: 709 KNWLSGSITKVVRNGRDTSFWSEKWSRG-----RQYSRLFKILLDKEAKVVDLR 855
>BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 789
Score = 37.0 bits (84), Expect = 0.014
Identities = 18/66 (27%), Positives = 36/66 (54%)
Frame = -3
Query: 2 RNAVSRNLFKGFKIKNDGTGISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKVN 61
+ A+ + G K+ + I+HL +ADDT+ G+++ + L +++ + ASG +N
Sbjct: 313 QQALRKGTLPGVKVARNCPPINHLLFADDTMFFGKSNASSCAILLSIMDKYRAASGRCIN 134
Query: 62 FYKSCL 67
KS +
Sbjct: 133 *TKSAI 116
>TC91262 similar to GP|20804797|dbj|BAB92481. putative amino acid or GABA
permease {Oryza sativa (japonica cultivar-group)},
partial (36%)
Length = 904
Score = 34.7 bits (78), Expect = 0.071
Identities = 17/38 (44%), Positives = 26/38 (67%), Gaps = 2/38 (5%)
Frame = +1
Query: 125 GNKYVSLG--GRIVLLNAVINSIPIFYLSFLKMPNKVW 160
G+KYV++ G I++L+ +INS+PI +LSFL W
Sbjct: 502 GSKYVTIAIHGGILVLHGIINSLPISWLSFLGQLAAFW 615
>TC82948
Length = 705
Score = 34.3 bits (77), Expect = 0.092
Identities = 15/44 (34%), Positives = 25/44 (56%)
Frame = +3
Query: 180 KLCWVKWRVVCQPRSKGGLGVRDIRVVNLSLLAKWRWRVLQGEE 223
K+ V W VC+P +G LG+R + +N +L K W ++ +E
Sbjct: 255 KVVKVSWEKVCRPIKEGSLGIRSLSKLNEALNLKLCWDMMISKE 386
>AW774658 similar to GP|2808681|emb| Hcr9-4B {Lycopersicon hirsutum}, partial
(4%)
Length = 665
Score = 27.7 bits (60), Expect(2) = 1.0
Identities = 13/22 (59%), Positives = 15/22 (68%)
Frame = -1
Query: 20 TGISHLQYADDTLCIGEASVDN 41
T SHLQ+ADDTL +G S N
Sbjct: 392 TVFSHLQFADDTLLLGVKSWAN 327
Score = 21.6 bits (44), Expect(2) = 1.0
Identities = 10/24 (41%), Positives = 16/24 (66%)
Frame = -3
Query: 45 LKALLRGFEMASGLKVNFYKSCLM 68
L+++L FE SGLKVN + ++
Sbjct: 321 LRSILVIFENMSGLKVNLREEVII 250
>BQ140834
Length = 560
Score = 29.6 bits (65), Expect = 2.3
Identities = 12/24 (50%), Positives = 18/24 (75%)
Frame = -1
Query: 367 KSCYGKLERLLGGEVEWSLEELRV 390
K YGKL++ +GGEV W + ++RV
Sbjct: 236 KRFYGKLKQNVGGEVGWKIIKIRV 165
>TC83174
Length = 1323
Score = 29.6 bits (65), Expect = 2.3
Identities = 12/24 (50%), Positives = 18/24 (75%)
Frame = -3
Query: 367 KSCYGKLERLLGGEVEWSLEELRV 390
K YGKL++ +GGEV W + ++RV
Sbjct: 256 KRFYGKLKQNVGGEVGWKIIKIRV 185
>TC77827 homologue to GP|15131692|emb|CAC48393. putative aminoaldehyde
dehydrogenase {Pisum sativum}, complete
Length = 1904
Score = 29.3 bits (64), Expect = 3.0
Identities = 22/77 (28%), Positives = 39/77 (50%)
Frame = +1
Query: 96 LGLPVGANSFKLVTWEPLLEQLSRKLHSWGNKYVSLGGRIVLLNAVINSIPIFYLSFLKM 155
+GLP+ A F L + LL +L RK +W N + + + I ++ + LSF+ +
Sbjct: 214 IGLPLLA-PFVLAIYALLLLRLLRKNQNWLNLKLLILVNHSMKRRGIWTMLLVVLSFMLI 390
Query: 156 PNKVWRKIVKIQCDFLW 172
K W + +++C F W
Sbjct: 391 LLKNWMQSKRLRCLFQW 441
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.141 0.472
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,048,772
Number of Sequences: 36976
Number of extensions: 328299
Number of successful extensions: 1458
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 1433
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1449
length of query: 420
length of database: 9,014,727
effective HSP length: 99
effective length of query: 321
effective length of database: 5,354,103
effective search space: 1718667063
effective search space used: 1718667063
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC122162.14


