

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122162.12 - phase: 0
(400 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
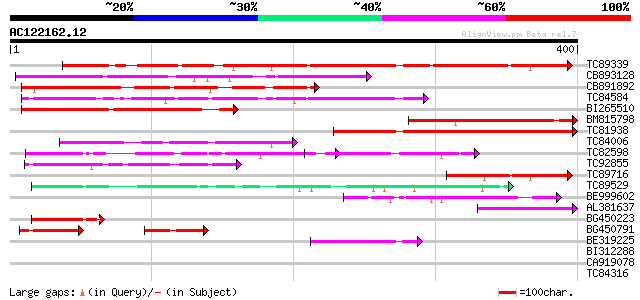
Score E
Sequences producing significant alignments: (bits) Value
TC89339 similar to GP|17380986|gb|AAL36305.1 unknown protein {Ar... 281 2e-76
CB893128 weakly similar to PIR|F96671|F966 hypothetical protein ... 197 5e-51
CB891892 183 9e-47
TC84584 similar to PIR|C84547|C84547 hypothetical protein At2g17... 166 1e-41
BI265510 weakly similar to PIR|C84555|C845 hypothetical protein ... 155 2e-38
BM815798 homologue to GP|7292702|gb|A CG15741 gene product {Dros... 143 1e-34
TC81938 similar to PIR|C84547|C84547 hypothetical protein At2g17... 136 2e-32
TC84006 homologue to GP|9758228|dbj|BAB08727.1 gb|AAD22308.1~gen... 123 1e-28
TC82598 weakly similar to PIR|C84547|C84547 hypothetical protein... 114 7e-26
TC92855 93 2e-19
TC89716 77 1e-14
TC89529 homologue to PIR|H86400|H86400 protein T17H3.7 [imported... 64 1e-10
BE999602 weakly similar to GP|6686405|gb|A F1E22.11 {Arabidopsis... 60 1e-09
AL381637 weakly similar to PIR|C96738|C967 unknown protein F3I17... 58 7e-09
BG450223 homologue to PIR|H71159|H711 hypothetical protein PH047... 50 2e-06
BG450791 weakly similar to GP|9758228|dbj| gb|AAD22308.1~gene_id... 50 2e-06
BE319225 47 1e-05
BI312288 similar to PIR|T49048|T490 hypothetical protein T5P19.1... 34 0.087
CA919078 34 0.11
TC84316 weakly similar to GP|11994612|dbj|BAB02749. gb|AAC24186.... 33 0.25
>TC89339 similar to GP|17380986|gb|AAL36305.1 unknown protein {Arabidopsis
thaliana}, partial (3%)
Length = 1290
Score = 281 bits (720), Expect = 2e-76
Identities = 168/368 (45%), Positives = 229/368 (61%), Gaps = 8/368 (2%)
Frame = +3
Query: 38 DLIRFRSVCSTWRSSPIPNDHNILPFQFPLLKYVPTPDSISNNNEIIDNIENTSTSFGHL 97
DLIRFRSVCSTWRSS +PN H ILP++FP+ K IS N+ + F HL
Sbjct: 3 DLIRFRSVCSTWRSSSVPNHHYILPYKFPVFKL----PFISEQND-------KNPPFCHL 149
Query: 98 SKRSFFLVKPPQEQQQQETLIRRRPWLIRITQNSSGKTQLLKPPFLSLTSTAFSHLPNV- 156
SK++ FL+KPPQ+QQ+Q+TL+R PWLIR++QN+ GKT+ P F S LP +
Sbjct: 150 SKQNIFLIKPPQQQQEQQTLLR--PWLIRVSQNTRGKTRFFHPLFCD------SPLPRIN 305
Query: 157 --VDFTKFSPQHLATDFIIDKDDLTFQNQ--HSSYLYPQKILAVTCPEKKPLVLGTLSYC 212
+DF K S HL ++ D Q+Q +S Y++P+K++AVTC K PL++ T+S
Sbjct: 306 LILDFRKLSLLHLGSNTFTRDFDTYSQHQPFNSDYMFPEKVIAVTCHGKNPLIVATIS-S 482
Query: 213 SSKRVLFHDRDKRWTPVSNLSTAHGDICLFKGRFYVVDQSGQTVTVGPDSSTELAAQPLY 272
+ +L +++W + ++S GDICLFKGR Y VD+ G+T+ VG DS+ L A+PL
Sbjct: 483 HPEPLLSKCGNEKWKVIPDMSMEFGDICLFKGRAYAVDKIGKTIMVGSDSNVHLVAEPLD 662
Query: 273 RRCVRGENRKLLVENEGELLLLDIHQTFFQFSIKFFKLDEMEKKWVKLKKLGGRVLFVGS 332
GEN+KLLVE++G+LLL +H F S+ FKL+E EKKWVKL LG +VLF+G
Sbjct: 663 G----GENKKLLVESDGDLLLACVH-AFPVISVDLFKLNEKEKKWVKLTSLGDKVLFLGI 827
Query: 333 GCSFSASGWDLCLPKGNCVIFIDTSVLSSDNMAG--GNRVFHLDRGQLSHVSEYPECENL 390
CSFSAS DL + KGNCVI I + + G + V LD+G L + + PE NL
Sbjct: 828 KCSFSASFSDLYVAKGNCVI-ISADIFRRLSRLGSQASYVLDLDQGWLPLLCDSPEYSNL 1004
Query: 391 FL-PPEWI 397
F PP+WI
Sbjct: 1005FWPPPKWI 1028
>CB893128 weakly similar to PIR|F96671|F966 hypothetical protein F13O11.14
[imported] - Arabidopsis thaliana, partial (8%)
Length = 768
Score = 197 bits (502), Expect = 5e-51
Identities = 125/264 (47%), Positives = 159/264 (59%), Gaps = 13/264 (4%)
Frame = +3
Query: 5 HTAVATSSMAS-VDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDH-NILP 62
H + +TS MA+ VDWSELPK+++ LISQ +D ELDLI FRS+CSTWRSS IPN H NI
Sbjct: 27 HHSASTSFMATEVDWSELPKELLNLISQCIDNELDLIHFRSICSTWRSSSIPNHHSNISS 206
Query: 63 FQFPLLKYVPTPDSISNN-NEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRR 121
F+FP KY+P P S N+ N+II I+N+++ F SKRS FLVK P Q + R
Sbjct: 207 FKFP--KYIPPPYSTKNDDNDIIHFIKNSNSPFDSFSKRSLFLVKAPPHQT-----VVLR 365
Query: 122 PWLIRITQ----NSSGKTQLL--KPPFLSLTSTAFSHL----PNVVDFTKFSPQHLATDF 171
PWL++I N GK L PP ++ +V+DF K S HL T +
Sbjct: 366 PWLMQICPSSLPNKRGKIMLTYSHPPIRHESAKQLQETLIERADVLDFNKLSVLHLGTRY 545
Query: 172 IIDKDDLTFQNQHSSYLYPQKILAVTCPEKKPLVLGTLSYCSSKRVLFHDRDKRWTPVSN 231
I NQ+ P+ LAVTC K PL+LGTLS+ S V+F +++W SN
Sbjct: 546 I--------SNQYGKC--PETALAVTCHGKNPLLLGTLSHFSHLPVIFRGCNEQWKLFSN 695
Query: 232 LSTAHGDICLFKGRFYVVDQSGQT 255
LST GDICLFKGRFYVV+ SG+T
Sbjct: 696 LSTVRGDICLFKGRFYVVEMSGRT 767
>CB891892
Length = 587
Score = 183 bits (465), Expect = 9e-47
Identities = 114/219 (52%), Positives = 136/219 (62%), Gaps = 9/219 (4%)
Frame = +2
Query: 9 ATSSMASV--DWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDH-NILPFQF 65
A SSMA DWS LP D++ LISQ +D E+DLIRFRSVC WRSS I H NILPF+F
Sbjct: 5 AASSMAPAMSDWSALPMDLVNLISQGIDTEIDLIRFRSVCCNWRSSSIRKHHLNILPFEF 184
Query: 66 PLLKYVPTPDSISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLI 125
PL K+ DSI N +NT F +LSK SFFL+KPP EQQQ TLIR RPWL+
Sbjct: 185 PLFKFRSLFDSIIN--------DNTIPFFCYLSKCSFFLIKPPHEQQQ--TLIRHRPWLV 334
Query: 126 RITQNSSGKTQLLKP------PFLSLTSTAFSHLPNVVDFTKFSPQHLATDFIIDKDDLT 179
RI Q S+GKTQ+L P PF SL S +DF K S HL TDFI+ D L
Sbjct: 335 RIRQYSNGKTQILNPLISYQSPFDSLHS---------IDFNKLSVIHLGTDFIVHDDSLL 487
Query: 180 FQNQHSSYLYPQKILAVTCPEKKPLVLGTLSYCSSKRVL 218
+ Y+YP+KI+A T EKKP+VLG + +SK +L
Sbjct: 488 Y-----DYMYPEKIVAFTGNEKKPIVLGAFT-TTSKPIL 586
>TC84584 similar to PIR|C84547|C84547 hypothetical protein At2g17030
[imported] - Arabidopsis thaliana, partial (4%)
Length = 812
Score = 166 bits (421), Expect = 1e-41
Identities = 115/293 (39%), Positives = 171/293 (58%), Gaps = 6/293 (2%)
Frame = +3
Query: 9 ATSSMASVDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNILPFQFPLL 68
A+SSMA DWS+LPKD++ LIS +LD E +RFRSVCS+WRSS N H+ L L
Sbjct: 18 ASSSMA--DWSQLPKDLLQLISSKLDSEFYQLRFRSVCSSWRSSVPKNHHHHL----TLP 179
Query: 69 KYVPTPDSISNNNEIIDNIENTSTSFGHLSKRSFFLVKPP--QEQQQQETLIRRRPWLIR 126
++PTP S++N N+ ++ LSKR+ FL+ PP QQ+TL PWLI+
Sbjct: 180 SHLPTP---SDSN----NLHHSKYITFPLSKRTIFLITPPTNHHHHQQQTL-NNNPWLIK 335
Query: 127 ITQNSSGKTQLLKPPFLSLTSTAFSHLPNVVDFTKFSPQHLATDFIIDKDDLTFQNQHSS 186
I +S +T+L P LS +LP V++F + L +F+I N++SS
Sbjct: 336 IGPDSRDRTRLWNP--LSRDKQLSIYLPQVINFNEHRVIDLGREFVIGN-----FNEYSS 494
Query: 187 YLYPQKILAVTCP----EKKPLVLGTLSYCSSKRVLFHDRDKRWTPVSNLSTAHGDICLF 242
LY +K++ + +++ VL T+ + S K LF D+RWT V + + D+C+F
Sbjct: 495 -LYMEKVIVLDADMWGGKERCSVLFTI-HISGKIALFRCGDERWTIVPEMPSPFDDVCVF 668
Query: 243 KGRFYVVDQSGQTVTVGPDSSTELAAQPLYRRCVRGENRKLLVENEGELLLLD 295
KGR VD +G+TV + PD S +L A+P++ G ++K LVE++GELLL+D
Sbjct: 669 KGRPVAVDGTGRTVALRPDLSLDLVAEPVF-----GGDKKFLVESDGELLLVD 812
>BI265510 weakly similar to PIR|C84555|C845 hypothetical protein At2g17690
[imported] - Arabidopsis thaliana, partial (8%)
Length = 545
Score = 155 bits (393), Expect = 2e-38
Identities = 85/155 (54%), Positives = 107/155 (68%), Gaps = 2/155 (1%)
Frame = +2
Query: 9 ATSSMASVDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNILPFQFPLL 68
A+SSMA DWS+LPKD++ LISQR+D E+DLIRFRS+CS WRSS IPN HNILPF+FPLL
Sbjct: 104 ASSSMAEADWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPNHHNILPFKFPLL 283
Query: 69 KYVPTPDSIS-NNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQ-QQQETLIRRRPWLIR 126
K +SI N+NEIID+I NT + F +LSKRS FLVKPP Q ++++TLI RRPW
Sbjct: 284 KC--EDNSIKYNDNEIIDSINNTDSLFCYLSKRSIFLVKPPPSQLEEKQTLIPRRPWFD* 457
Query: 127 ITQNSSGKTQLLKPPFLSLTSTAFSHLPNVVDFTK 161
I + + Q T+F+H N+ D K
Sbjct: 458 IYSKLTWQNQ-----------TSFTHSSNIPDHNK 529
>BM815798 homologue to GP|7292702|gb|A CG15741 gene product {Drosophila
melanogaster}, partial (14%)
Length = 577
Score = 143 bits (361), Expect = 1e-34
Identities = 77/132 (58%), Positives = 90/132 (67%), Gaps = 13/132 (9%)
Frame = +3
Query: 282 KLLVENEGELLLLDIHQTFFQFSIKFFKLDEM-------------EKKWVKLKKLGGRVL 328
K+LVE+EG LLLL I ++ FSI FF+LDE EKKWVKL+ G RV
Sbjct: 36 KMLVESEGRLLLLGIEESSNYFSIDFFELDEKKKKWVRLMDFDEKEKKWVKLRNFGDRVF 215
Query: 329 FVGSGCSFSASGWDLCLPKGNCVIFIDTSVLSSDNMAGGNRVFHLDRGQLSHVSEYPECE 388
F+G GCSFSAS DLC+ KGNC IFID SVL ++NM G RVFHLD+ +LS S+Y
Sbjct: 216 FIGRGCSFSASASDLCIQKGNCAIFIDESVLHNNNMVRGKRVFHLDQDRLSRGSKY---L 386
Query: 389 NLFLPPEWILKI 400
NLFLPPEWILKI
Sbjct: 387 NLFLPPEWILKI 422
>TC81938 similar to PIR|C84547|C84547 hypothetical protein At2g17030
[imported] - Arabidopsis thaliana, partial (1%)
Length = 916
Score = 136 bits (342), Expect = 2e-32
Identities = 78/176 (44%), Positives = 114/176 (64%), Gaps = 4/176 (2%)
Frame = +1
Query: 229 VSNLSTAHG-DICLFKGRFYVVDQSGQTVTVGPDSSTELAAQPLYRRCVRGENRKLLVEN 287
+ N+S +HG ++C+FKGR VVD++G+T +GPD +T L A+P++ G+++ L+ +
Sbjct: 1 IPNMSNSHGGNVCVFKGRPCVVDKTGRTFMIGPDLTTHLIAEPVFG----GKSKFLVESS 168
Query: 288 EGELLLLDIHQTF-FQFSIKFFKLDEMEKKWVKLKKLGGRVLFVGSGCSFSASGWDLCLP 346
E ELLL+D ++ + I +LDE EK+WVKL LG +VLF+G+G SFSAS +L
Sbjct: 169 EFELLLVDRYENYRVPVWIDVLRLDEKEKRWVKLANLGDKVLFLGNGYSFSASASELGFA 348
Query: 347 KGNCVIFIDTSVLSSDNMAGGN-RVFHLDRGQLSHVSEYPECENLFL-PPEWILKI 400
GNCVI+ + N+ VFHLD+GQ S +S+YPE LF PPEWI K+
Sbjct: 349 NGNCVIYSKSYGFHDLNIRKCKMSVFHLDQGQASPLSDYPEYFKLFWPPPEWIAKL 516
>TC84006 homologue to GP|9758228|dbj|BAB08727.1
gb|AAD22308.1~gene_id:MZF18.8~similar to unknown protein
{Arabidopsis thaliana}, partial (4%)
Length = 597
Score = 123 bits (308), Expect = 1e-28
Identities = 74/172 (43%), Positives = 98/172 (56%), Gaps = 4/172 (2%)
Frame = +2
Query: 36 ELDLIRFRSVCSTWRSSPIPNDHNILPFQFPLLKYVPTPDSISNNNEIIDNIENTSTSFG 95
E+DLI FRSVCSTWRSS +P H FQFPLLK+ PD I+ N SF
Sbjct: 137 EVDLIHFRSVCSTWRSSSVPKHHRNFRFQFPLLKFPFNPDFINRNK-----------SFC 283
Query: 96 HLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQNSSGKTQLLKPPFLSLTSTAFSHLPN 155
+++K++ FL+KPP QQE + RP LIRI QN+ GK++L P L L N
Sbjct: 284 YITKQNIFLIKPP----QQEQTLNLRPSLIRICQNARGKSKLYHP----LLQNGDKFLYN 439
Query: 156 -VVDFTKFSPQHLATDFIIDKDDLTFQNQ---HSSYLYPQKILAVTCPEKKP 203
+DF K S HL ++F + D TF ++ Y+YP+K++ VTC KKP
Sbjct: 440 FALDFNKLSILHLGSNFFMIDFDFTFNDKLYNPDYYMYPKKVVVVTCHGKKP 595
>TC82598 weakly similar to PIR|C84547|C84547 hypothetical protein At2g17030
[imported] - Arabidopsis thaliana, partial (2%)
Length = 1050
Score = 114 bits (285), Expect = 7e-26
Identities = 82/225 (36%), Positives = 117/225 (51%), Gaps = 3/225 (1%)
Frame = +1
Query: 12 SMA-SVDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNILPFQFPLLKY 70
SMA + DWS L KD++ LIS RLD E D+IRFRSVCSTWR IPN + LPF+ P
Sbjct: 55 SMAMAADWSTLHKDLLLLISSRLDNEADIIRFRSVCSTWRFF-IPN--HSLPFKLPHF-- 219
Query: 71 VPTPDSISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQN 130
+TS+ +LSK + FL+KPPQ++QQQ RPWLIRI +N
Sbjct: 220 ------------------HTSSLSCYLSKHNIFLIKPPQQEQQQTL----RPWLIRIAKN 333
Query: 131 SSGKTQLLKPPFLSLTSTAFSHLPNVVDFTKFSPQHLATDFIIDK--DDLTFQNQHSSYL 188
S +TQL P S+ S+ + H P+V+D KFS L D ++ + +D T + L
Sbjct: 334 SCRETQLFH-PLRSVDSSPY-HFPHVLDLKKFSVVRLGADVVMGRYNEDKTVLERSKELL 507
Query: 189 YPQKILAVTCPEKKPLVLGTLSYCSSKRVLFHDRDKRWTPVSNLS 233
++ + + P LG ++ H ++K + L+
Sbjct: 508 SQFELYQLKIHRQDPTPLGHGMGKKVVAIMQHGKNKETVALGTLN 642
Score = 75.9 bits (185), Expect = 3e-14
Identities = 52/130 (40%), Positives = 72/130 (55%), Gaps = 7/130 (5%)
Frame = +2
Query: 209 LSYCSSKRVLFHDRDKRWTPVSNLSTAHGDICLFKGRFYVVDQSGQTVTVGPDSSTELAA 268
+ YCS+ +K WT + +ST DIC+FK RFY V++ G+TV G D S EL A
Sbjct: 653 IQYCSTAS------EKHWTLIPCMSTYFLDICVFKERFYAVNKIGRTVAFGLDYSVELVA 814
Query: 269 QPLYRRCVRGENRKLLVENEGELLLLDIHQTF-FQF------SIKFFKLDEMEKKWVKLK 321
+ V G + K LVE EGELLL+DI + F F +K F+L+E +K+W + K
Sbjct: 815 E-----YVNGGDMKFLVECEGELLLVDICDSHCFGFPGEKGLKLKVFRLNERKKEW-RSK 976
Query: 322 KLGGRVLFVG 331
L R +G
Sbjct: 977 SLRNRGFVLG 1006
Score = 30.4 bits (67), Expect = 1.3
Identities = 16/29 (55%), Positives = 20/29 (68%), Gaps = 6/29 (20%)
Frame = +3
Query: 321 KKLGGR------VLFVGSGCSFSASGWDL 343
++ GGR VLF+G+GCSFSAS DL
Sbjct: 957 RRNGGRRA*GIGVLFLGNGCSFSASASDL 1043
>TC92855
Length = 625
Score = 93.2 bits (230), Expect = 2e-19
Identities = 66/155 (42%), Positives = 79/155 (50%), Gaps = 2/155 (1%)
Frame = +2
Query: 11 SSMASVDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPN--DHNILPFQFPLL 68
+SMA DWS LP +++ LISQ+LD L L+RFRSVC+TWRSS IPN N P P
Sbjct: 44 ASMA--DWSHLPSELLQLISQKLDTVLCLLRFRSVCATWRSSSIPNHKHKNSSPLNIP-- 211
Query: 69 KYVPTPDSISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRIT 128
P P+ N I NI + L + FL+KPPQ Q PW IRI
Sbjct: 212 -QFPEPNFDPFN---IRNIIKSKYITSRLITHTIFLIKPPQHQPS------LNPWWIRIG 361
Query: 129 QNSSGKTQLLKPPFLSLTSTAFSHLPNVVDFTKFS 163
+ GKTQL P S S NV+DF K S
Sbjct: 362 PDFKGKTQLWHP--FSSGQNLSSDFNNVLDFNKLS 460
>TC89716
Length = 735
Score = 77.0 bits (188), Expect = 1e-14
Identities = 48/95 (50%), Positives = 58/95 (60%), Gaps = 6/95 (6%)
Frame = +2
Query: 309 KLDEMEKKWVKLKKLGGRVLFVGSG--CSFSASGWDLCLPKGNCVIFIDTSVLSSDNMAG 366
KL+E EKKWVKL LG R+LF+G G CSFSA DLC+ KGN VIF + + S G
Sbjct: 2 KLNEKEKKWVKLTSLGDRILFLGLGLVCSFSACASDLCVAKGNSVIF-NNYIFESYRPFG 178
Query: 367 ---GNRVFHLDRGQLSHVSEYPECENLFLPP-EWI 397
V LD+G+ S +S+ PE NL PP EWI
Sbjct: 179 FQCKQYVLDLDQGRHSLLSDCPEYSNLLWPPAEWI 283
>TC89529 homologue to PIR|H86400|H86400 protein T17H3.7 [imported] -
Arabidopsis thaliana, partial (1%)
Length = 1745
Score = 63.5 bits (153), Expect = 1e-10
Identities = 93/365 (25%), Positives = 138/365 (37%), Gaps = 25/365 (6%)
Frame = +1
Query: 16 VDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNILPFQFPLLKYVPTPD 75
V+W+ELP++II IS+RL + D IRFR VC TW SS +P LP Q P+L
Sbjct: 448 VEWAELPQEIIESISKRLTIYADYIRFRCVCRTWNSS-VPKTPLHLPPQLPILML----- 609
Query: 76 SISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQNSSGKT 135
+ SF LS + P WL+ + + S+
Sbjct: 610 --------------SDESFFDLSTNKIHRLNFPLPSGPTRICGSSHGWLVILDETSN--L 741
Query: 136 QLLKPPFLSLTSTAFSHLPNVVDFTKFSPQHLATDFIIDKDDLTFQNQHSSYLYPQKILA 195
LL P + A LP++ F P + F +K++ LY K++
Sbjct: 742 NLLNP-----VTHATLSLPSLHTF----PYLVKKLFAFNKNN----------LYLIKVVL 864
Query: 196 VTCPEKKP-------LVLGTLSY----CSSKRVLFHDRDKRWTPVSNLSTAHGDICLFKG 244
+ P L L L++ C S +L + WT D G
Sbjct: 865 SSNPSPNDDFAAFAILGLRHLAFCRKGCDSWVLLDANVGDHWT----------DAVYKNG 1014
Query: 245 RFYVVDQSGQT----VTVGPDSS-----TELAAQPLYRRCVRGENRKLL---VENEGELL 292
F+ + SG T V GP S T L ++ GE L+ E E
Sbjct: 1015SFFAMSTSGITAVCDVVEGPRVSYIPPPTSLYYNDVFYAVFSGEEMLLIQRCCTEEYEPR 1194
Query: 293 LLDIHQTFFQFSIKFFKLDEMEKKWVKLKKLGGRVLFVGS--GCSFSASGWDLCLPKGNC 350
+ + +K++ KW +++ LG LF+G SFSA+ + C P NC
Sbjct: 1195DFEPYPDMSTVKFWIYKMNWNMLKWEEIQTLGEHSLFIGKNYSLSFSAADFAGCCP--NC 1368
Query: 351 VIFID 355
+ F D
Sbjct: 1369IYFTD 1383
>BE999602 weakly similar to GP|6686405|gb|A F1E22.11 {Arabidopsis thaliana},
partial (4%)
Length = 579
Score = 60.5 bits (145), Expect = 1e-09
Identities = 54/175 (30%), Positives = 85/175 (47%), Gaps = 21/175 (12%)
Frame = +1
Query: 236 HGDICLFKGRFYVVDQSGQTVTVGPDSSTELA--AQPLYRRCVRGENRKLLVENEGELLL 293
+ D+ +FKG+FYV D+ G T++ SS +L + PL C G N+K LVE+ G L +
Sbjct: 16 YDDLIVFKGQFYVTDKWG-TISWIDISSLKLIQFSPPL---CGFG-NKKHLVESCGNLYV 180
Query: 294 LDI-----------------HQTFFQF--SIKFFKLDEMEKKWVKLKKLGGRVLFVGSGC 334
+D HQ ++ K +KLDE KWV +K L R + C
Sbjct: 181 VDRYSYYEDENMGGNYDGRRHQARYEVVECFKVYKLDEEWGKWVDVKDLKNRAFILSPSC 360
Query: 335 SFSASGWDLCLPKGNCVIFIDTSVLSSDNMAGGNRVFHLDRGQLSHVSEYPECEN 389
+FS S +L +GNC+ F D+ + R+++LD +++ V P +N
Sbjct: 361 NFSVSAKELIGYQGNCIYFKDSFSV---------RMYNLDDCRITTVEFNPCIDN 498
>AL381637 weakly similar to PIR|C96738|C967 unknown protein F3I17.1
[imported] - Arabidopsis thaliana, partial (9%)
Length = 508
Score = 57.8 bits (138), Expect = 7e-09
Identities = 34/72 (47%), Positives = 43/72 (59%), Gaps = 2/72 (2%)
Frame = +3
Query: 331 GSGCSFSASGWDLCLPKGNCVIFIDTSVLSSDNMAGGNR-VFHLDRGQLSHVSEYPECEN 389
G+G SFSAS +L GNCVI+ + N+ VFHLD+GQ S +S+YPE
Sbjct: 3 GNGYSFSASASELGFANGNCVIYSKSYGFHDLNIRKCKMSVFHLDQGQASPLSDYPEYFK 182
Query: 390 LFL-PPEWILKI 400
LF PPEWI K+
Sbjct: 183 LFWPPPEWIAKL 218
>BG450223 homologue to PIR|H71159|H711 hypothetical protein PH0477 -
Pyrococcus horikoshii, partial (8%)
Length = 604
Score = 50.1 bits (118), Expect = 2e-06
Identities = 27/52 (51%), Positives = 35/52 (66%)
Frame = +1
Query: 16 VDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNILPFQFPL 67
VDWS LPK++ I + LD +D++RFRSVC ++RSS IP H P FPL
Sbjct: 274 VDWSHLPKELWSKIGKYLDNHIDILRFRSVCESFRSS-IPPSHPNSP-SFPL 423
>BG450791 weakly similar to GP|9758228|dbj|
gb|AAD22308.1~gene_id:MZF18.8~similar to unknown protein
{Arabidopsis thaliana}, partial (9%)
Length = 646
Score = 50.1 bits (118), Expect = 2e-06
Identities = 26/45 (57%), Positives = 34/45 (74%)
Frame = -2
Query: 8 VATSSMASVDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSS 52
+A S +A +WS+LP + + LISQ+L+ EL LIR RSVCS WRSS
Sbjct: 384 IAASMVA--EWSQLPHEFLQLISQKLNRELYLIRSRSVCSWWRSS 256
Score = 44.3 bits (103), Expect = 8e-05
Identities = 22/45 (48%), Positives = 28/45 (61%)
Frame = -1
Query: 96 HLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQNSSGKTQLLKP 140
+L K FL KPP+ QQ + +RPWLIRI + GKT+LL P
Sbjct: 160 YLIKHDIFLFKPPKHQQN----LHQRPWLIRIGPDFDGKTKLLHP 38
>BE319225
Length = 242
Score = 47.4 bits (111), Expect = 1e-05
Identities = 30/80 (37%), Positives = 47/80 (58%), Gaps = 1/80 (1%)
Frame = +2
Query: 213 SSKRVLFHDRDKRWTPVSNLSTAHGDICLFKGRFYVVDQSGQTVTVGPDSST-ELAAQPL 271
+SK +L ++ + ++ DIC+FKGR YVV ++G+T T+G D T EL A+ L
Sbjct: 11 TSKPILLKCSNENRNGIPVMTAWFEDICIFKGRPYVVHKTGRTATLGLDDFTFELVAESL 190
Query: 272 YRRCVRGENRKLLVENEGEL 291
G + K LVE++G+L
Sbjct: 191 VS---GGGDIKFLVESDGDL 241
>BI312288 similar to PIR|T49048|T490 hypothetical protein T5P19.120 -
Arabidopsis thaliana, partial (18%)
Length = 705
Score = 34.3 bits (77), Expect = 0.087
Identities = 22/66 (33%), Positives = 34/66 (51%)
Frame = +2
Query: 7 AVATSSMASVDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNILPFQFP 66
AV + ++ W++LP +++ L RLD+ D IR +VC W S + +L Q P
Sbjct: 35 AVKSDNLELQTWADLPAEVLELFLSRLDIG-DNIRASAVCKRWCS--VATSVRVLD-QSP 202
Query: 67 LLKYVP 72
L Y P
Sbjct: 203WLMYFP 220
>CA919078
Length = 515
Score = 33.9 bits (76), Expect = 0.11
Identities = 17/29 (58%), Positives = 20/29 (68%), Gaps = 1/29 (3%)
Frame = -1
Query: 370 VFHLDRGQLSHVSEYPECENLFL-PPEWI 397
V LD G+ S +S+YPE NLF PPEWI
Sbjct: 455 VLDLDLGRHSLLSDYPEYSNLFWPPPEWI 369
>TC84316 weakly similar to GP|11994612|dbj|BAB02749.
gb|AAC24186.1~gene_id:MGL6.4~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (10%)
Length = 892
Score = 32.7 bits (73), Expect = 0.25
Identities = 17/52 (32%), Positives = 31/52 (58%), Gaps = 4/52 (7%)
Frame = +3
Query: 21 LPKDIIFLISQRLDVELDLIRFRSVCSTWRS----SPIPNDHNILPFQFPLL 68
LP+++I +I RL V L+RF+ VC +W++ + N+H ++ +P L
Sbjct: 42 LPEELIVIILLRLPVR-SLLRFKCVCKSWKTLFSDTHFANNHFLISTVYPQL 194
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.138 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,834,448
Number of Sequences: 36976
Number of extensions: 240397
Number of successful extensions: 1315
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 1265
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1277
length of query: 400
length of database: 9,014,727
effective HSP length: 98
effective length of query: 302
effective length of database: 5,391,079
effective search space: 1628105858
effective search space used: 1628105858
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC122162.12


