

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122162.1 + phase: 0 /pseudo/partial
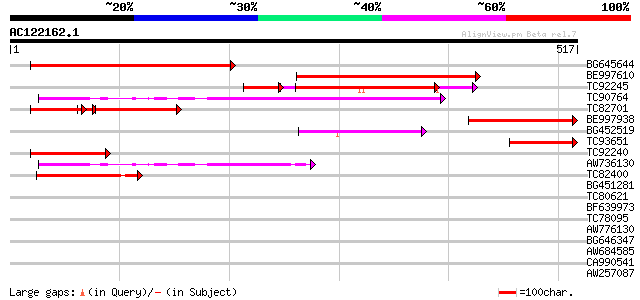
(517 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG645644 similar to GP|18958689|gb| putative membrane protein {O... 349 2e-96
BE997610 similar to GP|15912323|gb T8G24.8/T8G24.8 {Arabidopsis ... 323 1e-88
TC92245 weakly similar to GP|15912323|gb|AAL08295.1 T8G24.8/T8G2... 196 4e-61
TC90764 similar to PIR|T05666|T05666 hypothetical protein F22I13... 231 4e-61
TC82701 similar to GP|18958689|gb|AAL82672.1 putative membrane p... 152 1e-60
BE997938 similar to GP|15912323|gb| T8G24.8/T8G24.8 {Arabidopsis... 155 5e-38
BG452519 similar to PIR|D96551|D96 hypothetical protein F11M15.2... 129 2e-30
TC93651 similar to GP|15912323|gb|AAL08295.1 T8G24.8/T8G24.8 {Ar... 125 3e-29
TC92240 weakly similar to GP|18958689|gb|AAL82672.1 putative mem... 117 1e-26
AW736130 similar to PIR|T05666|T05 hypothetical protein F22I13.1... 111 8e-25
TC82400 similar to PIR|T02512|T02512 hypothetical protein At2g38... 104 7e-23
BG451281 similar to PIR|T06063|T060 hypothetical protein F19H22.... 35 0.070
TC80621 similar to GP|11994223|dbj|BAB01345. gb|AAF35421.1~gene_... 33 0.35
BF639973 similar to GP|17065002|gb Unknown protein {Arabidopsis ... 32 0.45
TC78095 weakly similar to PIR|G84776|G84776 hypothetical protein... 31 1.3
AW776130 similar to GP|17065002|gb| Unknown protein {Arabidopsis... 31 1.3
BG646347 similar to PIR|T05135|T05 hypothetical protein F7H19.22... 30 2.2
AW684585 similar to PIR|T05135|T051 hypothetical protein F7H19.2... 30 2.2
CA990541 weakly similar to GP|9758600|dbj gene_id:MDF20.10~pir||... 30 2.9
AW257087 homologue to GP|6683126|dbj KIAA0339 protein {Homo sapi... 30 2.9
>BG645644 similar to GP|18958689|gb| putative membrane protein {Oryza sativa
(japonica cultivar-group)}, partial (17%)
Length = 799
Score = 349 bits (895), Expect = 2e-96
Identities = 185/187 (98%), Positives = 186/187 (98%)
Frame = +3
Query: 20 ECSLVFKMDSLAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQ 79
+ SLVFKMDSLAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQ
Sbjct: 237 DASLVFKMDSLAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQ 416
Query: 80 ASRITIFPLVSITTSFVAEEDTMDRINTKAAEKQFNESGKAKSNEVMPDDHLLQDIEAGA 139
ASRITIFPLVSITTSFVAEEDTMDRINTKAAEKQFNESGKAKSNEVMPDDHLLQDIEAGA
Sbjct: 417 ASRITIFPLVSITTSFVAEEDTMDRINTKAAEKQFNESGKAKSNEVMPDDHLLQDIEAGA 596
Query: 140 TKQDSTLKNGDDANSNISKSSIVTNSSNKSESKPIRKKRHIASASTALLFGTVLGLIQAA 199
TKQDSTLKNGDDANSNISKSSIVTNSSNKSESKPIRKKRHIASASTALLFGTVLGLIQAA
Sbjct: 597 TKQDSTLKNGDDANSNISKSSIVTNSSNKSESKPIRKKRHIASASTALLFGTVLGLIQAA 776
Query: 200 TLIFAAK 206
TLIFAAK
Sbjct: 777 TLIFAAK 797
>BE997610 similar to GP|15912323|gb T8G24.8/T8G24.8 {Arabidopsis thaliana},
partial (30%)
Length = 512
Score = 323 bits (827), Expect = 1e-88
Identities = 168/168 (100%), Positives = 168/168 (100%)
Frame = +2
Query: 262 VSGYALNVAMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKVDLLPPSMKDLQI 321
VSGYALNVAMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKVDLLPPSMKDLQI
Sbjct: 8 VSGYALNVAMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKVDLLPPSMKDLQI 187
Query: 322 FRFLKNGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAVAI 381
FRFLKNGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAVAI
Sbjct: 188 FRFLKNGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAVAI 367
Query: 382 QAILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGV 429
QAILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGV
Sbjct: 368 QAILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGV 511
>TC92245 weakly similar to GP|15912323|gb|AAL08295.1 T8G24.8/T8G24.8
{Arabidopsis thaliana}, partial (29%)
Length = 640
Score = 196 bits (499), Expect(2) = 4e-61
Identities = 98/132 (74%), Positives = 116/132 (87%)
Frame = +3
Query: 261 IVSGYALNVAMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKVDLLPPSMKDLQ 320
+ +GY+LNV ++PLLIF K+GI+GAAI+HV+SQY+MA L FILMKKV LLPP +KDLQ
Sbjct: 144 LXAGYSLNVLLEPLLIFKLKMGIKGAAIAHVISQYMMAFTLFFILMKKVYLLPPRIKDLQ 323
Query: 321 IFRFLKNGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAVA 380
IFRFL+NGGLL+ +VIAVTFCVTL+ASLAARLG IPMAAFQ CLQVW+ SSL ADGLA+A
Sbjct: 324 IFRFLRNGGLLMTKVIAVTFCVTLAASLAARLGSIPMAAFQPCLQVWLASSLFADGLAIA 503
Query: 381 IQAILACSFAEK 392
+QAILA SFA K
Sbjct: 504 VQAILAGSFAXK 539
Score = 57.4 bits (137), Expect = 1e-08
Identities = 62/194 (31%), Positives = 82/194 (41%), Gaps = 15/194 (7%)
Frame = +2
Query: 248 FRGFKDTTTPLYVIVSGYALNVAMDPLLIFYFKLGI-RGAAISHVLSQYIMATLLLFILM 306
F GFKDTTTPLYVI G + + + F + G R + + LS + LL I
Sbjct: 104 FXGFKDTTTPLYVIXCGIFIECVIGAITYF*IENGHKRSSNCTCYLSVHDGIYPLLHINE 283
Query: 307 KKVDLLPPSMK-----DLQI---FRFLKNGGLLLARVIAVTFCVTLSASLAARLGPIPMA 358
K V P+ K D Q+ +R + + G + +++ G
Sbjct: 284 KSVS---PTSKNKGSSDFQVS*KWRSVDDKGN----------SSNILCDFSSKFGCKVRF 424
Query: 359 AFQTCLQVWMTSSLLADGLAVAIQAILACS------FAEKDYNKVTTAATRTLQMSFVLG 412
CL TS L L +C+ F D NK T AATRTLQ F+LG
Sbjct: 425 NSHGCLSTLPTS--LVSILPFC*WFGYSCTGNPSWFFRGXDCNKTTAAATRTLQFXFILG 598
Query: 413 VGLSLVVGGGLYFG 426
GLS + G GLYFG
Sbjct: 599 AGLSXIXGFGLYFG 640
Score = 57.0 bits (136), Expect(2) = 4e-61
Identities = 28/36 (77%), Positives = 31/36 (85%)
Frame = +1
Query: 214 LKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFR 249
+K+ SPML PAVKYL R+ GAPAVLLSLAMQGIFR
Sbjct: 1 VKHGSPMLKPAVKYLTYRSFGAPAVLLSLAMQGIFR 108
>TC90764 similar to PIR|T05666|T05666 hypothetical protein F22I13.150 -
Arabidopsis thaliana, partial (57%)
Length = 1219
Score = 231 bits (590), Expect = 4e-61
Identities = 147/371 (39%), Positives = 204/371 (54%)
Frame = +2
Query: 27 MDSLAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRITIF 86
+ + +EI+ ++ P+ A DPIA L++TA+IG LG +ELA++GVS + IF
Sbjct: 275 ISDVKREIISLSLPALAGQAIDPIAQLMETAYIGRLGTLELASSGVS---------VVIF 427
Query: 87 PLVSITTSFVAEEDTMDRINTKAAEKQFNESGKAKSNEVMPDDHLLQDIEAGATKQDSTL 146
++S K FN +P L + QD
Sbjct: 428 NIIS---------------------KLFN----------IP----LLSVATSFVAQDM-- 496
Query: 147 KNGDDANSNISKSSIVTNSSNKSESKPIRKKRHIASASTALLFGTVLGLIQAATLIFAAK 206
+NIS S N K + S STALL +G+ +A L F ++
Sbjct: 497 -------ANISSSQNANNPQRKQ----------LPSVSTALLLALGIGIFEALALYFGSR 625
Query: 207 PLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIVSGYA 266
L +G+ +P LVPA K+L LRA GAPAV+LSLA+QGIFRGFKDT TP+ + G
Sbjct: 626 MFLRLIGVAAVNPTLVPAQKFLSLRAFGAPAVVLSLALQGIFRGFKDTKTPVICLGIGNL 805
Query: 267 LNVAMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKVDLLPPSMKDLQIFRFLK 326
V + PLL++YFKLG+ GAAIS VLSQYI L+++ L K+ LLPP M +LQ ++K
Sbjct: 806 SAVFLFPLLMYYFKLGVAGAAISTVLSQYIGTLLMIWCLNKRAVLLPPKMGNLQFGGYIK 985
Query: 327 NGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAVAIQAILA 386
+GG +L R +AV +TL S+AAR GP+ MAA Q C+QVW+ SLL D LAV+ QA++A
Sbjct: 986 SGGFVLGRTLAVLTTMTLGTSMAARHGPVAMAAHQICMQVWLAVSLLTDALAVSGQALIA 1165
Query: 387 CSFAEKDYNKV 397
S + +Y V
Sbjct: 1166SSLSRHEYKAV 1198
>TC82701 similar to GP|18958689|gb|AAL82672.1 putative membrane protein
{Oryza sativa (japonica cultivar-group)}, partial (17%)
Length = 670
Score = 152 bits (385), Expect(3) = 1e-60
Identities = 77/78 (98%), Positives = 78/78 (99%)
Frame = +2
Query: 79 QASRITIFPLVSITTSFVAEEDTMDRINTKAAEKQFNESGKAKSNEVMPDDHLLQDIEAG 138
+ASRITIFPLVSITTSFVAEEDTMDRINTKAAEKQFNESGKAKSNEVMPDDHLLQDIEAG
Sbjct: 437 KASRITIFPLVSITTSFVAEEDTMDRINTKAAEKQFNESGKAKSNEVMPDDHLLQDIEAG 616
Query: 139 ATKQDSTLKNGDDANSNI 156
ATKQDSTLKNGDDANSNI
Sbjct: 617 ATKQDSTLKNGDDANSNI 670
Score = 92.4 bits (228), Expect(3) = 1e-60
Identities = 47/51 (92%), Positives = 49/51 (95%)
Frame = +3
Query: 20 ECSLVFKMDSLAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAA 70
+ SLVFKMDSLAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELA +
Sbjct: 258 DASLVFKMDSLAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAGS 410
Score = 27.7 bits (60), Expect(3) = 1e-60
Identities = 13/18 (72%), Positives = 15/18 (83%)
Frame = +1
Query: 63 GPVELAAAGVSIAVFNQA 80
G + L AAGVSIAVFNQ+
Sbjct: 388 GRLNLQAAGVSIAVFNQS 441
>BE997938 similar to GP|15912323|gb| T8G24.8/T8G24.8 {Arabidopsis thaliana},
partial (18%)
Length = 537
Score = 155 bits (391), Expect = 5e-38
Identities = 75/99 (75%), Positives = 85/99 (85%)
Frame = -3
Query: 419 VGGGLYFGAGVFSKNVAVIHLIRLGLPFVAATQPINSLAFVFDGVNYGASDFAYSAYSLV 478
VG GLYFGAG+FSKN+ VIH IR+G P VAATQPIN+LAFVFDGVNYGASDFAY++YSLV
Sbjct: 535 VGFGLYFGAGIFSKNLQVIHFIRIGAPIVAATQPINTLAFVFDGVNYGASDFAYASYSLV 356
Query: 479 MVSIASVTSLFFLYKSKGFIGIWIALTIYMSLRMFAGVW 517
VS+ SV F LY+S FIGIWIAL+IYM+LRM AGVW
Sbjct: 355 TVSLVSVGVEFLLYRSNQFIGIWIALSIYMTLRMLAGVW 239
>BG452519 similar to PIR|D96551|D96 hypothetical protein F11M15.20 [imported]
- Arabidopsis thaliana, partial (23%)
Length = 643
Score = 129 bits (325), Expect = 2e-30
Identities = 76/167 (45%), Positives = 93/167 (55%), Gaps = 50/167 (29%)
Frame = +1
Query: 264 GYALNVAMDPLLIFYFKLGIRGAAISHVLSQYIM-------------------------- 297
G N+A+DPL IF F++G+ GAAI+HV+SQY++
Sbjct: 1 GDLTNIALDPLFIFVFRMGVNGAAIAHVISQYVLLGCDSIL*L*FNRDLRNTLCRAILSL 180
Query: 298 ------------------------ATLLLFILMKKVDLLPPSMKDLQIFRFLKNGGLLLA 333
+ +LL+ L K+VDL+PPS+K +Q RF KNG LL
Sbjct: 181 NFLLLLFNFVISHFQTNCKCRYLLSAILLWSLNKQVDLIPPSIKHMQFDRFAKNGFLLFM 360
Query: 334 RVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAVA 380
RVIAVTFCVTLSASLAA G MAAFQ CLQVW+ SLLADGLAVA
Sbjct: 361 RVIAVTFCVTLSASLAAHHGSTSMAAFQVCLQVWLAVSLLADGLAVA 501
>TC93651 similar to GP|15912323|gb|AAL08295.1 T8G24.8/T8G24.8 {Arabidopsis
thaliana}, partial (12%)
Length = 686
Score = 125 bits (315), Expect = 3e-29
Identities = 62/62 (100%), Positives = 62/62 (100%)
Frame = +2
Query: 456 LAFVFDGVNYGASDFAYSAYSLVMVSIASVTSLFFLYKSKGFIGIWIALTIYMSLRMFAG 515
LAFVFDGVNYGASDFAYSAYSLVMVSIASVTSLFFLYKSKGFIGIWIALTIYMSLRMFAG
Sbjct: 2 LAFVFDGVNYGASDFAYSAYSLVMVSIASVTSLFFLYKSKGFIGIWIALTIYMSLRMFAG 181
Query: 516 VW 517
VW
Sbjct: 182 VW 187
>TC92240 weakly similar to GP|18958689|gb|AAL82672.1 putative membrane
protein {Oryza sativa (japonica cultivar-group)},
partial (15%)
Length = 576
Score = 117 bits (292), Expect = 1e-26
Identities = 59/73 (80%), Positives = 66/73 (89%)
Frame = +2
Query: 20 ECSLVFKMDSLAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQ 79
+ L+FK+D+L+KEILGIA PSAL AADPIASLIDTAFIGHLGPVELAAAGVSIA+FNQ
Sbjct: 242 DARLIFKLDALSKEILGIAIPSALTEAADPIASLIDTAFIGHLGPVELAAAGVSIALFNQ 421
Query: 80 ASRITIFPLVSIT 92
S+ITIFPL SIT
Sbjct: 422 XSKITIFPLXSIT 460
>AW736130 similar to PIR|T05666|T05 hypothetical protein F22I13.150 -
Arabidopsis thaliana, partial (31%)
Length = 692
Score = 111 bits (277), Expect = 8e-25
Identities = 88/253 (34%), Positives = 121/253 (47%)
Frame = +1
Query: 27 MDSLAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRITIF 86
+ + +EI+ ++ P+ A DPIA L++TA+IG LG +ELA++GVS + IF
Sbjct: 85 ISDVKREIISLSLPALAGQAIDPIAQLMETAYIGRLGTLELASSGVS---------VVIF 237
Query: 87 PLVSITTSFVAEEDTMDRINTKAAEKQFNESGKAKSNEVMPDDHLLQDIEAGATKQDSTL 146
++S K FN +P L + QD
Sbjct: 238 NIIS---------------------KLFN----------IP----LLSVATSFVAQDM-- 306
Query: 147 KNGDDANSNISKSSIVTNSSNKSESKPIRKKRHIASASTALLFGTVLGLIQAATLIFAAK 206
+NIS S N K + S STALL +G+ +A L F ++
Sbjct: 307 -------ANISSSQNANNPQRKQ----------LPSVSTALLLALGIGIFEALALYFGSR 435
Query: 207 PLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIVSGYA 266
L +G+ +P LVPA K+L LRA GAPAV+LSLA+QGIFRGFKDT TP VI G
Sbjct: 436 MFLRLIGVAAVNPTLVPAQKFLSLRAFGAPAVVLSLALQGIFRGFKDTKTP--VICLGKF 609
Query: 267 LNVAMDPLLIFYF 279
L +L+FYF
Sbjct: 610 LVYC---VLLFYF 639
>TC82400 similar to PIR|T02512|T02512 hypothetical protein At2g38330
[imported] - Arabidopsis thaliana, partial (13%)
Length = 589
Score = 104 bits (260), Expect = 7e-23
Identities = 54/97 (55%), Positives = 72/97 (73%)
Frame = +3
Query: 25 FKMDSLAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRIT 84
FK D L ++L IA P+A+A+AADPIASLIDTAF+GH+G VELAA GVS +VFN S++
Sbjct: 294 FKFDELGMDMLSIALPAAVALAADPIASLIDTAFVGHIGAVELAAVGVSASVFNLVSKVF 473
Query: 85 IFPLVSITTSFVAEEDTMDRINTKAAEKQFNESGKAK 121
PL++ITTSFVAEE + I + +Q E+GK++
Sbjct: 474 NVPLLNITTSFVAEEQAL--IGKEEESEQAEENGKSE 578
>BG451281 similar to PIR|T06063|T060 hypothetical protein F19H22.130 -
Arabidopsis thaliana, partial (12%)
Length = 556
Score = 35.0 bits (79), Expect = 0.070
Identities = 20/54 (37%), Positives = 29/54 (53%)
Frame = +1
Query: 32 KEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRITI 85
KEI+ P+ P+ SLIDTA IG +ELAA G + V + S +++
Sbjct: 394 KEIVKFTAPATGLWICGPLMSLIDTAVIGQGSSIELAALGPATVVCDYMSYVSM 555
>TC80621 similar to GP|11994223|dbj|BAB01345.
gb|AAF35421.1~gene_id:MJK13.21~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (22%)
Length = 1883
Score = 32.7 bits (73), Expect = 0.35
Identities = 22/83 (26%), Positives = 38/83 (45%), Gaps = 5/83 (6%)
Frame = -3
Query: 99 EDTMDRINTKAAEKQFNESGKAKS---NEVMPDDHLLQDIEAGATKQDSTLKNGDDAN-- 153
ED + ++N K +K+ + K N + + +LQD + K+D ++N DDA
Sbjct: 801 EDIIQQLNQKFPQKELLKVSKVSDSDINNIEKELQVLQDKKCWTEKEDDKIENVDDATGY 622
Query: 154 SNISKSSIVTNSSNKSESKPIRK 176
N S + V ++ E I K
Sbjct: 621 KNYSAEAYVGETNGSFEKTKIGK 553
>BF639973 similar to GP|17065002|gb Unknown protein {Arabidopsis thaliana},
partial (22%)
Length = 679
Score = 32.3 bits (72), Expect = 0.45
Identities = 22/63 (34%), Positives = 32/63 (49%)
Frame = +2
Query: 32 KEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRITIFPLVSI 91
KEI+ P+ P+ SLIDTA +G +ELAA G + + + +F +SI
Sbjct: 323 KEIVLFTGPAIGLWLCGPLMSLIDTAVVGQGSSIELAALGPATVFCDYLGYLFMF--LSI 496
Query: 92 TTS 94
TS
Sbjct: 497 ATS 505
>TC78095 weakly similar to PIR|G84776|G84776 hypothetical protein At2g36090
[imported] - Arabidopsis thaliana, partial (19%)
Length = 1467
Score = 30.8 bits (68), Expect = 1.3
Identities = 18/63 (28%), Positives = 30/63 (47%), Gaps = 5/63 (7%)
Frame = +2
Query: 155 NISKSSIVTN-----SSNKSESKPIRKKRHIASASTALLFGTVLGLIQAATLIFAAKPLL 209
N+S S I+ S N S P+ +RH + +L+GTV+G++Q + +
Sbjct: 749 NLSLSWIIIEPTRKRSVNLSSRLPVTVRRHWLTGELEVLYGTVMGMVQCMVKVTCCGKVG 928
Query: 210 GAM 212
G M
Sbjct: 929 GEM 937
>AW776130 similar to GP|17065002|gb| Unknown protein {Arabidopsis thaliana},
partial (17%)
Length = 571
Score = 30.8 bits (68), Expect = 1.3
Identities = 17/40 (42%), Positives = 22/40 (54%)
Frame = +1
Query: 32 KEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAG 71
KEI+ P+ P+ SLIDTA +G +ELAA G
Sbjct: 289 KEIVLFTGPAIGLWLCGPLMSLIDTAVVGQGSSIELAALG 408
>BG646347 similar to PIR|T05135|T05 hypothetical protein F7H19.220 -
Arabidopsis thaliana, partial (33%)
Length = 752
Score = 30.0 bits (66), Expect = 2.2
Identities = 23/67 (34%), Positives = 33/67 (48%)
Frame = +3
Query: 10 QNANSMFY*FECSLVFKMDSLAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAA 69
+N FY +M+S++K IAFP L S+I F+GHL +LA
Sbjct: 162 KNQTFSFYHHLTHTFKEMNSISK----IAFPMILTCLLLYCRSMISMLFLGHLN--DLAL 323
Query: 70 AGVSIAV 76
AG S+A+
Sbjct: 324 AGGSLAI 344
>AW684585 similar to PIR|T05135|T051 hypothetical protein F7H19.220 -
Arabidopsis thaliana, partial (15%)
Length = 458
Score = 30.0 bits (66), Expect = 2.2
Identities = 21/48 (43%), Positives = 26/48 (53%), Gaps = 3/48 (6%)
Frame = +3
Query: 32 KEILGI---AFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAV 76
KE++ I AFP S+I F+GHLG ELA AG S+AV
Sbjct: 231 KEVISISKIAFPMIFTGLLLYCRSMISMLFLGHLG--ELALAGGSLAV 368
>CA990541 weakly similar to GP|9758600|dbj
gene_id:MDF20.10~pir||T08929~similar to unknown protein
{Arabidopsis thaliana}, partial (13%)
Length = 712
Score = 29.6 bits (65), Expect = 2.9
Identities = 22/97 (22%), Positives = 39/97 (39%), Gaps = 1/97 (1%)
Frame = +2
Query: 91 ITTSFVAEEDTMDRINTKAAEKQFNESGKAKSNEVMPDDHLLQDIEAGATKQDSTLKNGD 150
+T S + E + A++ SG S ++D K+ +T +
Sbjct: 239 VTKSVLLAEQEKSAKEKRIAKQGSTGSGTPTSRRSAKSRKKIEDSSIAEKKKTATDTESE 418
Query: 151 DANSNISKSSIVTNSSNKSES-KPIRKKRHIASASTA 186
+K + S+KSE KPI+ K+HIA ++
Sbjct: 419 SEKDEENKEENEIDVSDKSEDEKPIKGKKHIAKQGSS 529
>AW257087 homologue to GP|6683126|dbj KIAA0339 protein {Homo sapiens},
partial (2%)
Length = 466
Score = 29.6 bits (65), Expect = 2.9
Identities = 17/55 (30%), Positives = 31/55 (55%), Gaps = 3/55 (5%)
Frame = +1
Query: 134 DIEAGATKQDSTLKNGDDANSNISKSSIVTNSSNKSE---SKPIRKKRHIASAST 185
D ++ ++ S+ + ++S+ S SS +NS ++ E +KP+ KK SAST
Sbjct: 292 DSDSSSSSSSSSSSSSSSSSSSSSSSSSSSNSDDEEEEEEAKPVVKKTPAKSAST 456
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.327 0.139 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,797,336
Number of Sequences: 36976
Number of extensions: 158351
Number of successful extensions: 1080
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 1067
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1080
length of query: 517
length of database: 9,014,727
effective HSP length: 100
effective length of query: 417
effective length of database: 5,317,127
effective search space: 2217241959
effective search space used: 2217241959
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC122162.1


