

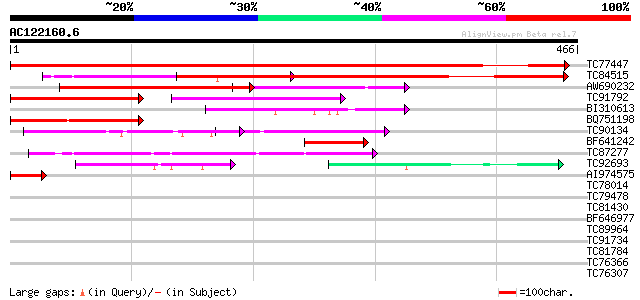
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122160.6 + phase: 0
(466 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77447 similar to GP|16974501|gb|AAL31160.1 AT3g06720/F3E22_14 ... 598 e-171
TC84515 similar to GP|14595713|gb|AAK70912.1 putative impotin al... 425 e-119
AW690232 similar to GP|13752562|gb importin alpha 2 {Capsicum an... 322 3e-88
TC91792 homologue to GP|13752562|gb|AAK38727.1 importin alpha 2 ... 177 6e-45
BI310613 similar to GP|13752562|gb importin alpha 2 {Capsicum an... 153 1e-37
BQ751198 homologue to GP|12718253|em probable importin alpha SRP... 122 4e-28
TC90134 similar to PIR|T48328|T48328 importin alpha-like protein... 97 1e-20
BF641242 similar to GP|14595713|gb| putative impotin alpha 1b {O... 67 1e-11
TC87277 similar to PIR|T45588|T45588 arm repeat containing prote... 61 8e-10
TC92693 similar to PIR|T48328|T48328 importin alpha-like protein... 56 3e-08
AI974575 similar to GP|13752562|gb importin alpha 2 {Capsicum an... 47 2e-05
TC78014 similar to GP|17529090|gb|AAL38755.1 unknown protein {Ar... 40 0.002
TC79478 similar to GP|18491179|gb|AAL69492.1 unknown protein {Ar... 40 0.002
TC81430 weakly similar to GP|18491179|gb|AAL69492.1 unknown prot... 40 0.002
BF646977 weakly similar to PIR|T06629|T06 hypothetical protein T... 35 0.047
TC89964 weakly similar to PIR|B96639|B96639 protein T1F9.16 [imp... 35 0.061
TC91734 weakly similar to GP|16604661|gb|AAL24123.1 unknown prot... 33 0.18
TC81784 similar to PIR|T40294|T40294 hypothetical protein SPBC35... 33 0.18
TC76366 homologue to SP|O65194|RBS_MEDSA Ribulose bisphosphate c... 33 0.30
TC76307 homologue to SP|O65194|RBS_MEDSA Ribulose bisphosphate c... 33 0.30
>TC77447 similar to GP|16974501|gb|AAL31160.1 AT3g06720/F3E22_14
{Arabidopsis thaliana}, partial (96%)
Length = 2271
Score = 598 bits (1542), Expect = e-171
Identities = 306/462 (66%), Positives = 358/462 (77%), Gaps = 2/462 (0%)
Frame = +2
Query: 1 MVADIWSDDNSQQLEATTEFRKRLSVDK-PPIDDVIQSGVVPRFVQFLDKGDFPQLQLEA 59
+V IW+DDN+ Q EATT+FRK LS+++ PPI++VIQ+GVVPRF++FL + DFPQLQ EA
Sbjct: 338 LVTGIWTDDNNMQFEATTQFRKLLSIERSPPIEEVIQAGVVPRFIEFLMREDFPQLQFEA 517
Query: 60 AWALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPRGRDLV 119
AWALTNIA+GTSENTKVV++ GAVP+FVKLL+SP DDVR QA WALGN+AGDSPR RDLV
Sbjct: 518 AWALTNIASGTSENTKVVIEAGAVPIFVKLLASPSDDVREQAVWALGNVAGDSPRCRDLV 697
Query: 120 LSHGALILLLSQLNEQEGLYRLRNAVWTLSNFCRGKPQPALEQVRSALPALKCLVFSKDE 179
L HGAL+ LL+QLNE L LRNA WTLSNFCRGKPQPA +QV+ ALPAL L+ S DE
Sbjct: 698 LGHGALLPLLAQLNEHAKLSMLRNATWTLSNFCRGKPQPAFDQVKPALPALASLIHSNDE 877
Query: 180 VVLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGDD 239
VLT+ACWALSYLSDG +D IQ VIEAGVC RLV++LL PS S L PALRTVGNIV+GDD
Sbjct: 878 EVLTDACWALSYLSDGANDKIQGVIEAGVCSRLVELLLHPSPSVLIPALRTVGNIVTGDD 1057
Query: 240 MQTQAIVNHGLLPCLLSLLTHP-KKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVN 298
+QTQ I+NH +LP L +LL + KKSIKKEAC T+SNITAGN++QIQ VI+A +IAPLVN
Sbjct: 1058LQTQVIINHQVLPPLTNLLINSYKKSIKKEACWTISNITAGNKQQIQDVIDANIIAPLVN 1237
Query: 299 LLQNAEFDITKEVAQALTNVTSGGTHEHIKYLVSQGCIKPLCDLLVCPDPRIVTVCLEGL 358
LLQNAEFDI KE A A++N TSGG+HE +KYLVSQGCIKPLCDLL CPDPRIVTVCLEGL
Sbjct: 1238LLQNAEFDIKKEAAWAISNATSGGSHEQLKYLVSQGCIKPLCDLLTCPDPRIVTVCLEGL 1417
Query: 359 ENFLKVGEAEKSISNTGDVNLYAQTIKDVEGLEKGLEKFLKVGEAEKRFGNTGDVNLYAQ 418
EN LKVGEA+K+ T VN YA+ I + EG
Sbjct: 1418ENILKVGEADKNAGITDGVNRYAELIDEAEG----------------------------- 1510
Query: 419 MIEDVKGLEKIENLDSHEDREIYEKAAKILERYWLEDEDETL 460
LEKIENL SH++ EIYEKA KILE YWLE++DE +
Sbjct: 1511-------LEKIENLQSHDNNEIYEKAVKILETYWLEEDDEAM 1615
>TC84515 similar to GP|14595713|gb|AAK70912.1 putative impotin alpha 1b
{Oryza sativa}, partial (56%)
Length = 979
Score = 425 bits (1093), Expect = e-119
Identities = 219/323 (67%), Positives = 246/323 (75%), Gaps = 1/323 (0%)
Frame = +3
Query: 138 LYRLRNAVWTLSNFCRGKPQPALEQVRSALPALKCLVFSKDEVVLTEACWALSYLSDGTS 197
L LRNA WTLSNFCRGKPQP +QV+ ALPAL L+ S DE VLT+ACWALSYLSDGT+
Sbjct: 9 LSMLRNATWTLSNFCRGKPQPPFDQVKPALPALAGLIHSNDEEVLTDACWALSYLSDGTN 188
Query: 198 DNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGDDMQTQAIVNHGLLPCLLSL 257
D IQAVIEAGVC RLV++LL PS S L PALRTVGNIV+GDD+QTQ I+NH LPCL +L
Sbjct: 189 DKIQAVIEAGVCPRLVELLLHPSPSVLIPALRTVGNIVTGDDLQTQVIINHQALPCLYNL 368
Query: 258 LTHP-KKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVNLLQNAEFDITKEVAQALT 316
LT+ KKSIKKEAC T+SNITAGN++QIQAVIEA + PLV+LLQNAEFDI KE A A++
Sbjct: 369 LTNNYKKSIKKEACWTISNITAGNKQQIQAVIEANIFGPLVSLLQNAEFDIKKEAAWAIS 548
Query: 317 NVTSGGTHEHIKYLVSQGCIKPLCDLLVCPDPRIVTVCLEGLENFLKVGEAEKSISNTGD 376
N TSGG+HE IKYLVSQGCIKPLCDLL+CPDPRIVTVCLEGLEN
Sbjct: 549 NATSGGSHEQIKYLVSQGCIKPLCDLLICPDPRIVTVCLEGLENI--------------- 683
Query: 377 VNLYAQTIKDVEGLEKGLEKFLKVGEAEKRFGNTGDVNLYAQMIEDVKGLEKIENLDSHE 436
LKVGEA+K GN GDVNLYAQMI+D +GLEKIENL SH+
Sbjct: 684 ---------------------LKVGEADKNIGNNGDVNLYAQMIDDAEGLEKIENLQSHD 800
Query: 437 DREIYEKAAKILERYWLEDEDET 459
+ EIYEKA KILE YWLE+EDET
Sbjct: 801 NTEIYEKAVKILETYWLEEEDET 869
Score = 70.1 bits (170), Expect = 2e-12
Identities = 55/214 (25%), Positives = 97/214 (44%), Gaps = 6/214 (2%)
Frame = +3
Query: 28 KPPIDDVIQSGVVPRFVQFLDKGDFPQLQLEAAWALTNIAAGTSENTKVVVDHGAVPMFV 87
+PP D V +P + D ++ +A WAL+ ++ GT++ + V++ G P V
Sbjct: 66 QPPFDQV--KPALPALAGLIHSND-EEVLTDACWALSYLSDGTNDKIQAVIEAGVCPRLV 236
Query: 88 KLLSSPCDDVRGQAAWALGNIAGDSPRGRDLVLSHGALILLLSQLNEQEGLYRLRNAVWT 147
+LL P V A +GNI ++++H AL L + L + A WT
Sbjct: 237 ELLLHPSPSVLIPALRTVGNIVTGDDLQTQVIINHQALPCLYNLLTNNYKKSIKKEACWT 416
Query: 148 LSNFCRGKPQPALEQVRSALPA-----LKCLVFSKDEVVLTEACWALS-YLSDGTSDNIQ 201
+SN G Q Q+++ + A L L+ + + + EA WA+S S G+ + I+
Sbjct: 417 ISNITAGNKQ----QIQAVIEANIFGPLVSLLQNAEFDIKKEAAWAISNATSGGSHEQIK 584
Query: 202 AVIEAGVCGRLVQILLEPSLSALFPALRTVGNIV 235
++ G L +L+ P + L + NI+
Sbjct: 585 YLVSQGCIKPLCDLLICPDPRIVTVCLEGLENIL 686
>AW690232 similar to GP|13752562|gb importin alpha 2 {Capsicum annuum},
partial (30%)
Length = 481
Score = 322 bits (824), Expect = 3e-88
Identities = 159/160 (99%), Positives = 159/160 (99%)
Frame = +2
Query: 42 RFVQFLDKGDFPQLQLEAAWALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQA 101
RFVQFLDKGDFPQLQLEAAWALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQA
Sbjct: 2 RFVQFLDKGDFPQLQLEAAWALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQA 181
Query: 102 AWALGNIAGDSPRGRDLVLSHGALILLLSQLNEQEGLYRLRNAVWTLSNFCRGKPQPALE 161
AWALGNIAGDSPRGRDLVLSHGALILLLSQLNEQEGLYRLRNAVWTLSNFCRGKPQPALE
Sbjct: 182 AWALGNIAGDSPRGRDLVLSHGALILLLSQLNEQEGLYRLRNAVWTLSNFCRGKPQPALE 361
Query: 162 QVRSALPALKCLVFSKDEVVLTEACWALSYLSDGTSDNIQ 201
QVRS LPALKCLVFSKDEVVLTEACWALSYLSDGTSDNIQ
Sbjct: 362 QVRSXLPALKCLVFSKDEVVLTEACWALSYLSDGTSDNIQ 481
Score = 52.4 bits (124), Expect = 4e-07
Identities = 39/146 (26%), Positives = 67/146 (45%), Gaps = 1/146 (0%)
Frame = +2
Query: 184 EACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGDDMQTQ 243
EA WAL+ ++ GTS+N + V++ G V++L P A +GNI
Sbjct: 50 EAAWALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPRGRD 229
Query: 244 AIVNHGLLPCLLSLLTHPKKSIK-KEACRTLSNITAGNREQIQAVIEAGLIAPLVNLLQN 302
+++HG L LLS L + + + A TLSN G + + + L P + L
Sbjct: 230 LVLSHGALILLLSQLNEQEGLYRLRNAVWTLSNFCRGKPQPALEQVRSXL--PALKCLVF 403
Query: 303 AEFDITKEVAQALTNVTSGGTHEHIK 328
++ ++ A + S GT ++I+
Sbjct: 404 SKDEVVLTEACWALSYLSDGTSDNIQ 481
>TC91792 homologue to GP|13752562|gb|AAK38727.1 importin alpha 2 {Capsicum
annuum}, partial (34%)
Length = 655
Score = 177 bits (450), Expect = 6e-45
Identities = 86/111 (77%), Positives = 100/111 (89%), Gaps = 1/111 (0%)
Frame = +2
Query: 1 MVADIWSDDNSQQLEATTEFRKRLSVDK-PPIDDVIQSGVVPRFVQFLDKGDFPQLQLEA 59
MVA +WSDDN+QQLEATT++RK LS+++ PPI++VIQSGVVPRFVQ L + DFPQLQ EA
Sbjct: 323 MVAGVWSDDNNQQLEATTQYRKLLSIERSPPIEEVIQSGVVPRFVQLLVREDFPQLQSEA 502
Query: 60 AWALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAG 110
AWALTNIA+GTSENTKVV+DHGAVP+FVKLL SP DDVR QA WALGN+AG
Sbjct: 503 AWALTNIASGTSENTKVVIDHGAVPIFVKLLXSPSDDVREQAVWALGNVAG 655
Score = 54.7 bits (130), Expect = 7e-08
Identities = 41/145 (28%), Positives = 67/145 (45%), Gaps = 2/145 (1%)
Frame = +2
Query: 134 EQEGLYRLRNAVWTLSNFCRGKPQPALEQVRSALPALKCLVFSKDEVVLTEACWAL-SYL 192
E+ L + R + F + +E+ +LPA+ V+S D EA L
Sbjct: 215 EESLLKKRREGLQNQPQFTPLQSSSIVEKK*ESLPAMVAGVWSDDNNQQLEATTQYRKLL 394
Query: 193 SDGTSDNIQAVIEAGVCGRLVQILLEPSLSAL-FPALRTVGNIVSGDDMQTQAIVNHGLL 251
S S I+ VI++GV R VQ+L+ L A + NI SG T+ +++HG +
Sbjct: 395 SIERSPPIEEVIQSGVVPRFVQLLVREDFPQLQSEAAWALTNIASGTSENTKVVIDHGAV 574
Query: 252 PCLLSLLTHPKKSIKKEACRTLSNI 276
P + LL P ++++A L N+
Sbjct: 575 PIFVKLLXSPSDDVREQAVWALGNV 649
>BI310613 similar to GP|13752562|gb importin alpha 2 {Capsicum annuum},
partial (28%)
Length = 725
Score = 153 bits (387), Expect = 1e-37
Identities = 102/193 (52%), Positives = 117/193 (59%), Gaps = 26/193 (13%)
Frame = -3
Query: 162 QVRSALPALKCLVFSKDEVVLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQILL---- 217
Q+R ALPALK LVFSKDE VLT+A WALSYLSDGT+DNIQAVIEAGVCGRLVQ+LL
Sbjct: 615 QMRPALPALKHLVFSKDEEVLTDAWWALSYLSDGTNDNIQAVIEAGVCGRLVQLLL*VPI 436
Query: 218 ------EPSLSALFPALRTVGNIVSGDDMQTQAIVNHG--LLPCLLSLLTHP-------- 261
L L + G + + G L LLS++ H
Sbjct: 435 LIPWSANILLQLLIILALNICACFEGTHLLQC*FLKFGGTLFVRLLSIMVHSHAF*AC*P 256
Query: 262 ---KKSIKKE---ACRTLSNITAGNREQIQAVIEAGLIAPLVNLLQNAEFDITKEVAQAL 315
+K ++K+ C+TL I +QAVIEAGLIAPLVNLLQNAEFDI KE A AL
Sbjct: 255 VVTRKVLRKKFAGLCQTLQLICT-----VQAVIEAGLIAPLVNLLQNAEFDIKKEAAWAL 91
Query: 316 TNVTSGGTHEHIK 328
TN TSGGTHE IK
Sbjct: 90 TNATSGGTHEQIK 52
Score = 33.1 bits (74), Expect = 0.23
Identities = 18/46 (39%), Positives = 27/46 (58%), Gaps = 1/46 (2%)
Frame = -3
Query: 31 IDDVIQSGVVPRFVQFLDKGDFPQLQLEAAWALTN-IAAGTSENTK 75
+ VI++G++ V L +F ++ EAAWALTN + GT E K
Sbjct: 186 VQAVIEAGLIAPLVNLLQNAEF-DIKKEAAWALTNATSGGTHEQIK 52
>BQ751198 homologue to GP|12718253|em probable importin alpha SRP1
{Neurospora crassa}, partial (35%)
Length = 813
Score = 122 bits (305), Expect = 4e-28
Identities = 64/111 (57%), Positives = 86/111 (76%), Gaps = 1/111 (0%)
Frame = +3
Query: 1 MVADIWSDDNSQQLEATTEFRKRLSVDK-PPIDDVIQSGVVPRFVQFLDKGDFPQLQLEA 59
MV ++SD Q++ATT+FRK LS ++ PPI++VI++GVV RFV+FL + +Q EA
Sbjct: 483 MVQGVFSDQIDLQIQATTKFRKLLSKERNPPIEEVIKTGVVSRFVEFL-RSPHTLVQFEA 659
Query: 60 AWALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAG 110
AWALTNIA+G++ T+VV++ GAVP+FV+LL SP DVR QA WALGNIAG
Sbjct: 660 AWALTNIASGSATQTQVVIEAGAVPIFVELLGSPEPDVREQAVWALGNIAG 812
Score = 37.4 bits (85), Expect = 0.012
Identities = 21/59 (35%), Positives = 31/59 (51%)
Frame = +3
Query: 176 SKDEVVLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNI 234
S +V EA WAL+ ++ G++ Q VIEAG V++L P A+ +GNI
Sbjct: 630 SPHTLVQFEAAWALTNIASGSATQTQVVIEAGAVPIFVELLGSPEPDVREQAVWALGNI 806
>TC90134 similar to PIR|T48328|T48328 importin alpha-like protein -
Arabidopsis thaliana, partial (50%)
Length = 763
Score = 97.4 bits (241), Expect = 1e-20
Identities = 72/193 (37%), Positives = 103/193 (53%), Gaps = 11/193 (5%)
Frame = +2
Query: 12 QQLEATTEFRKRLSVDK-PPIDDVIQSGVVPRFVQFLDKGDFPQLQLEAAWALTNIAAGT 70
+++ A E R+ LS + PP++ +++G V VQ L G + LEAAW LTNIAAG
Sbjct: 158 KRVGALQELRRLLSRSEFPPVESAVKAGAVAILVQCLSFGSPDEQLLEAAWCLTNIAAGN 337
Query: 71 SENTKVVVDHGAVPMFVKLL----SSPCDDVRGQAAWALGNIAGDSPRGRDLVLSHGALI 126
E TK ++ A+P+ + L SSP V Q AWALGN+AG+ R+++L GAL+
Sbjct: 338 PEETKALLP--ALPLLIAHLGEKSSSP---VAEQCAWALGNVAGEDEELRNVLLIQGALV 502
Query: 127 LLLSQLNEQEGLYR---LRNAVWTLSNFCRG-KPQPALEQVR--SALPALKCLVFSKDEV 180
L + YR +R A W LSN +G P+ A E +R L A+ + D+
Sbjct: 503 PLARMMLP----YRRSTVRTAAWALSNLIKGPNPKAANELIRVDGVLDAIVRHLTKADDE 670
Query: 181 VLTEACWALSYLS 193
TE W + YLS
Sbjct: 671 SATEVAWVVVYLS 709
Score = 41.2 bits (95), Expect = 9e-04
Identities = 37/146 (25%), Positives = 67/146 (45%), Gaps = 3/146 (2%)
Frame = +2
Query: 170 LKCLVFSKDEVVLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALR 229
++CL F + L EA W L+ ++ G + +A++ A L+ L E S S +
Sbjct: 257 VQCLSFGSPDEQLLEAAWCLTNIAAGNPEETKALLPA--LPLLIAHLGEKSSSPVAEQCA 430
Query: 230 -TVGNIVSGDDMQTQAIVNHGLLPCLLSLLTHPKKSIKKEACRTLSNITAG-NREQIQAV 287
+GN+ D+ ++ G L L ++ ++S + A LSN+ G N + +
Sbjct: 431 WALGNVAGEDEELRNVLLIQGALVPLARMMLPYRRSTVRTAAWALSNLIKGPNPKAANEL 610
Query: 288 IEA-GLIAPLVNLLQNAEFDITKEVA 312
I G++ +V L A+ + EVA
Sbjct: 611 IRVDGVLDAIVRHLTKADDESATEVA 688
>BF641242 similar to GP|14595713|gb| putative impotin alpha 1b {Oryza
sativa}, partial (11%)
Length = 680
Score = 67.0 bits (162), Expect = 1e-11
Identities = 35/54 (64%), Positives = 44/54 (80%), Gaps = 1/54 (1%)
Frame = +1
Query: 243 QAIVNHGLLPCLLSLLTHP-KKSIKKEACRTLSNITAGNREQIQAVIEAGLIAP 295
Q I+NH +LP L +LL + KKSIKKEAC T+SNITAGN++QIQ VI+A +IAP
Sbjct: 517 QVIINHQVLPPLTNLLINSYKKSIKKEACWTISNITAGNKQQIQDVIDAHIIAP 678
Score = 31.6 bits (70), Expect = 0.68
Identities = 14/41 (34%), Positives = 22/41 (53%)
Frame = +1
Query: 168 PALKCLVFSKDEVVLTEACWALSYLSDGTSDNIQAVIEAGV 208
P L+ S + + EACW +S ++ G IQ VI+A +
Sbjct: 547 PLTNLLINSYKKSIKKEACWTISNITAGNKQQIQDVIDAHI 669
Score = 29.3 bits (64), Expect = 3.4
Identities = 13/46 (28%), Positives = 22/46 (47%)
Frame = +1
Query: 34 VIQSGVVPRFVQFLDKGDFPQLQLEAAWALTNIAAGTSENTKVVVD 79
+I V+P L ++ EA W ++NI AG + + V+D
Sbjct: 523 IINHQVLPPLTNLLINSYKKSIKKEACWTISNITAGNKQQIQDVID 660
>TC87277 similar to PIR|T45588|T45588 arm repeat containing protein homolog
- Arabidopsis thaliana, partial (48%)
Length = 1393
Score = 61.2 bits (147), Expect = 8e-10
Identities = 76/288 (26%), Positives = 130/288 (44%), Gaps = 1/288 (0%)
Frame = +1
Query: 16 ATTEFRKRLSVDKPPIDDVIQSGVVPRFVQFLDKGDFPQLQLEAAWALTNIAAGTSENTK 75
+T+E K S P I+S +Q L G P+ Q AA + +A ++N
Sbjct: 94 STSEPSKSASACTPAERSKIES-----LIQKLTSGG-PEDQRSAAGEIRLLAKRNADNRV 255
Query: 76 VVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIA-GDSPRGRDLVLSHGALILLLSQLNE 134
+ + GA+P+ V LLS P + A AL N++ +S +G ++S GA+ ++ L +
Sbjct: 256 AIAEAGAIPLLVGLLSVPDSRTQEHAVTALLNLSIYESNKGS--IVSSGAVPGIVHVL-K 426
Query: 135 QEGLYRLRNAVWTLSNFCRGKPQPALEQVRSALPALKCLVFSKDEVVLTEACWALSYLSD 194
+ + NA TL + A+ L L+ + +A AL L
Sbjct: 427 RGSMEARENAAATLFSLSVIDENKVTIGSSGAISPLVTLLSEGTQRGKKDAATALFNLCI 606
Query: 195 GTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGDDMQTQAIVNHGLLPCL 254
+ +AV AGV L+++L EPS + AL + + S + + AI +P L
Sbjct: 607 YQGNKGKAV-RAGVIPTLMRLLTEPSGGMVDEALAILAILASHPEGKA-AIGAAEAVPVL 780
Query: 255 LSLLTHPKKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVNLLQN 302
+ + + K+ A L ++ +G+R+ I E G++APL+ L QN
Sbjct: 781 VKFIGNGSPRNKENAAAVLVHLCSGDRQYIAQAQELGVMAPLLELAQN 924
>TC92693 similar to PIR|T48328|T48328 importin alpha-like protein -
Arabidopsis thaliana, partial (32%)
Length = 770
Score = 55.8 bits (133), Expect = 3e-08
Identities = 48/202 (23%), Positives = 80/202 (38%), Gaps = 9/202 (4%)
Frame = +2
Query: 263 KSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVNLLQNAEFDITKEVAQALTNVTSGG 322
+ ++KEA LSNI AG+ E Q + + + L++L A FDI KEVA L N+
Sbjct: 68 QGLRKEAAWVLSNIAAGSMEHKQLIYSSETLPLLLHLFSAAPFDIRKEVAYILGNICVAP 247
Query: 323 TH---------EHIKYLVSQGCIKPLCDLLVCPDPRIVTVCLEGLENFLKVGEAEKSISN 373
T EH+ LV +GC+ DL+ D + L+ +E L+
Sbjct: 248 TKGDEIPNLILEHLVSLVEKGCLPGFIDLVRSADIEAAKLGLQFIELVLR---------- 397
Query: 374 TGDVNLYAQTIKDVEGLEKGLEKFLKVGEAEKRFGNTGDVNLYAQMIEDVKGLEKIENLD 433
G+ G +++E G+E +E
Sbjct: 398 ---------------GMPNGKG---------------------PKLVEQEDGIEAMERFQ 469
Query: 434 SHEDREIYEKAAKILERYWLED 455
HE+ ++ A ++++Y+ ED
Sbjct: 470 FHENEDVRTMANSLVDKYFGED 535
Score = 50.8 bits (120), Expect = 1e-06
Identities = 40/145 (27%), Positives = 70/145 (47%), Gaps = 14/145 (9%)
Frame = +2
Query: 55 LQLEAAWALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPR 114
L+ EAAW L+NIAAG+ E+ +++ +P+ + L S+ D+R + A+ LGNI +
Sbjct: 74 LRKEAAWVLSNIAAGSMEHKQLIYSSETLPLLLHLFSAAPFDIRKEVAYILGNICVAPTK 253
Query: 115 GRD---LVLSHGALILLLSQ--------LNEQEGLYRLRNAVWTLSNFCRGKPQ---PAL 160
G + L+L H L+ L+ + L + + + + RG P P L
Sbjct: 254 GDEIPNLILEH--LVSLVEKGCLPGFIDLVRSADIEAAKLGLQFIELVLRGMPNGKGPKL 427
Query: 161 EQVRSALPALKCLVFSKDEVVLTEA 185
+ + A++ F ++E V T A
Sbjct: 428 VEQEDGIEAMERFQFHENEDVRTMA 502
>AI974575 similar to GP|13752562|gb importin alpha 2 {Capsicum annuum},
partial (19%)
Length = 392
Score = 46.6 bits (109), Expect = 2e-05
Identities = 19/30 (63%), Positives = 26/30 (86%)
Frame = +1
Query: 1 MVADIWSDDNSQQLEATTEFRKRLSVDKPP 30
MVA +WSDDN+ QLE+TT+FRK LS+++ P
Sbjct: 295 MVAGVWSDDNNLQLESTTQFRKLLSIERTP 384
>TC78014 similar to GP|17529090|gb|AAL38755.1 unknown protein {Arabidopsis
thaliana}, partial (74%)
Length = 2305
Score = 39.7 bits (91), Expect = 0.002
Identities = 60/257 (23%), Positives = 108/257 (41%), Gaps = 1/257 (0%)
Frame = +3
Query: 47 LDKGDFPQLQLEAAWALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALG 106
L D Q Q AA L +A ++N + + GA+P+ V LLSS + A AL
Sbjct: 1299 LTSNDIEQ-QRAAAGELRLLAKRNADNRVCIAEAGAIPLLVDLLSSTDPRTQEHADTALL 1475
Query: 107 NIA-GDSPRGRDLVLSHGALILLLSQLNEQEGLYRLRNAVWTLSNFCRGKPQPALEQVRS 165
N++ +S +G +++ GA+ ++ L + + NA TL +
Sbjct: 1476 NLSINESNKG--TIVNAGAIPDIVDVL-KNGSMEARENAAATLFSLSVLDENKVAIGAAG 1646
Query: 166 ALPALKCLVFSKDEVVLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALF 225
A+PAL L+ +A A+ L + +AV +AG+ L++ + + +
Sbjct: 1647 AIPALIKLLCEGTPRGKKDAATAIFNLCIYQGNKARAV-KAGIVAPLIRFMKDAGGGMVD 1823
Query: 226 PALRTVGNIVSGDDMQTQAIVNHGLLPCLLSLLTHPKKSIKKEACRTLSNITAGNREQIQ 285
AL + I++G AI +P L+ ++ ++ A L + G+ Q++
Sbjct: 1824 EAL-AILTILAGHHEGRTAIGQAEPIPILVEVIRTGSPRNRENAAAVLWLVCTGDLLQLK 2000
Query: 286 AVIEAGLIAPLVNLLQN 302
E G L L +N
Sbjct: 2001 LAKEHGAEEALQGLSEN 2051
Score = 39.3 bits (90), Expect = 0.003
Identities = 39/155 (25%), Positives = 69/155 (44%)
Frame = +3
Query: 164 RSALPALKCLVFSKDEVVLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSA 223
++A+ AL + S D A L L+ +DN + EAG LV +L
Sbjct: 1269 KTAIKALLDKLTSNDIEQQRAAAGELRLLAKRNADNRVCIAEAGAIPLLVDLLSSTDPRT 1448
Query: 224 LFPALRTVGNIVSGDDMQTQAIVNHGLLPCLLSLLTHPKKSIKKEACRTLSNITAGNREQ 283
A + N+ S ++ IVN G +P ++ +L + ++ A TL +++ + +
Sbjct: 1449 QEHADTALLNL-SINESNKGTIVNAGAIPDIVDVLKNGSMEARENAAATLFSLSVLDENK 1625
Query: 284 IQAVIEAGLIAPLVNLLQNAEFDITKEVAQALTNV 318
+ A+ AG I L+ LL K+ A A+ N+
Sbjct: 1626 V-AIGAAGAIPALIKLLCEGTPRGKKDAATAIFNL 1727
>TC79478 similar to GP|18491179|gb|AAL69492.1 unknown protein {Arabidopsis
thaliana}, partial (39%)
Length = 958
Score = 39.7 bits (91), Expect = 0.002
Identities = 62/286 (21%), Positives = 104/286 (35%), Gaps = 8/286 (2%)
Frame = +2
Query: 54 QLQLEAAWALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSP 113
++Q AA AL +A EN +V+ A+P + +L S + +A +GN+ SP
Sbjct: 86 KVQRAAAGALRTLAFKNDENKNQIVECNALPTLILMLRSEDAAIHYEAVGVIGNLVHSSP 265
Query: 114 RGRDLVLSHGALILLLSQLNEQEGLYRLRNAVWTLSNFCRGKPQPALEQVRSALPALKCL 173
+ V+ GA L+ + LS+ C +
Sbjct: 266 NIKKDVILAGA----------------LQPVIGLLSSCCSESQR---------------- 349
Query: 174 VFSKDEVVLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGN 233
EA L + SD +++ G L+++L + + +G
Sbjct: 350 ----------EAALLLGQFAATDSDCKVHIVQRGAVRPLIEMLQSSDVQLKEMSAFALGR 499
Query: 234 IVSGDDMQTQAIVNH--GLLPCLLSLLTHPKKSIKKEACRTLSNITAGNREQIQAVIEAG 291
+ D QA + H GL+P LL LL S++ A L + A N + + I G
Sbjct: 500 L--AQDTHNQAGIAHSGGLVP-LLKLLDSKNGSLQHNAAFALYGL-AENEDNVPDFIRIG 667
Query: 292 LIAPLVNLLQNAEF------DITKEVAQALTNVTSGGTHEHIKYLV 331
I Q+ EF D + + L +G H+ YL+
Sbjct: 668 GI----KRFQDGEFIIQATKDCVAKTLKRLEEKINGRVLNHLLYLM 793
Score = 39.3 bits (90), Expect = 0.003
Identities = 35/116 (30%), Positives = 56/116 (48%)
Frame = +2
Query: 33 DVIQSGVVPRFVQFLDKGDFPQLQLEAAWALTNIAAGTSENTKVVVDHGAVPMFVKLLSS 92
DVI +G + + L + Q EAA L AA S+ +V GAV +++L S
Sbjct: 278 DVILAGALQPVIGLLSSC-CSESQREAALLLGQFAATDSDCKVHIVQRGAVRPLIEMLQS 454
Query: 93 PCDDVRGQAAWALGNIAGDSPRGRDLVLSHGALILLLSQLNEQEGLYRLRNAVWTL 148
++ +A+ALG +A D+ + S G L+ LL L+ + G + NA + L
Sbjct: 455 SDVQLKEMSAFALGRLAQDTHNQAGIAHS-GGLVPLLKLLDSKNGSLQ-HNAAFAL 616
>TC81430 weakly similar to GP|18491179|gb|AAL69492.1 unknown protein
{Arabidopsis thaliana}, partial (49%)
Length = 1491
Score = 39.7 bits (91), Expect = 0.002
Identities = 28/82 (34%), Positives = 46/82 (55%), Gaps = 1/82 (1%)
Frame = +2
Query: 68 AGTSENTKV-VVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPRGRDLVLSHGALI 126
A T + KV +V GAV +++LSSP +R +A+ALG +A D+ + + +G L+
Sbjct: 20 AATDSDCKVHIVQRGAVRPLIEMLSSPDVQLREMSAFALGRLAQDT-HNQAGIAHNGGLV 196
Query: 127 LLLSQLNEQEGLYRLRNAVWTL 148
LL L+ + G + NA + L
Sbjct: 197 PLLKLLDSKNGSLQ-HNAAFAL 259
>BF646977 weakly similar to PIR|T06629|T06 hypothetical protein T20K18.60 -
Arabidopsis thaliana, partial (29%)
Length = 662
Score = 35.4 bits (80), Expect = 0.047
Identities = 40/163 (24%), Positives = 73/163 (44%), Gaps = 2/163 (1%)
Frame = +1
Query: 56 QLEAAWALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPRG 115
++EAA + + +S+ + D G + + +LSS + R + AL N+A + R
Sbjct: 133 KIEAAREIRRMVRKSSKTRSKLADSGVIQPLIFMLSSSNIEARESSLLALLNLAVRNERN 312
Query: 116 RDLVLSHGALILLLSQLNEQ-EGLYRLRN-AVWTLSNFCRGKPQPALEQVRSALPALKCL 173
+ +++ GA+ L+ L Q G+ L A+ TLS+ KP + A P L +
Sbjct: 313 KVQIVTAGAVPPLVELLKMQSNGIRELATAAILTLSSAAPNKP---IIAASGAAPLLVQI 483
Query: 174 VFSKDEVVLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQIL 216
+ S + L LS T + I+ + +A L+ +L
Sbjct: 484 LKSGSVQGKVDTVTTLHNLSYSTVNPIE-LXDASAVSPLINLL 609
Score = 28.5 bits (62), Expect = 5.7
Identities = 28/117 (23%), Positives = 50/117 (41%)
Frame = +1
Query: 188 ALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGDDMQTQAIVN 247
AL L+ N ++ AG LV++L S A + + S + I
Sbjct: 277 ALLNLAVRNERNKVQIVTAGAVPPLVELLKMQSNGIRELATAAILTLSSAAPNKP-IIAA 453
Query: 248 HGLLPCLLSLLTHPKKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVNLLQNAE 304
G P L+ +L K + TL N++ I+ + +A ++PL+NLL++ +
Sbjct: 454 SGAAPLLVQILKSGSVQGKVDTVTTLHNLSYSTVNPIE-LXDASAVSPLINLLKDCK 621
>TC89964 weakly similar to PIR|B96639|B96639 protein T1F9.16 [imported] -
Arabidopsis thaliana, partial (35%)
Length = 928
Score = 35.0 bits (79), Expect = 0.061
Identities = 31/103 (30%), Positives = 50/103 (48%), Gaps = 3/103 (2%)
Frame = +1
Query: 38 GVVPRFVQFLDKGD--FPQLQLEAAWALTNIAAGTSENTKVVV-DHGAVPMFVKLLSSPC 94
G V + + ++ G+ +L L+ A+ L+ GTSE K + D G + FVK L++
Sbjct: 256 GFVDQLLYYVRHGEVSIQELALKVAFRLS----GTSEEAKKAMGDAGFMVEFVKFLNAKS 423
Query: 95 DDVRGQAAWALGNIAGDSPRGRDLVLSHGALILLLSQLNEQEG 137
+VR AA AL + + V + LLL L+ +EG
Sbjct: 424 FEVREMAAEALSGMVTVPRNRKRFVQDDHNIALLLQLLDPEEG 552
>TC91734 weakly similar to GP|16604661|gb|AAL24123.1 unknown protein
{Arabidopsis thaliana}, partial (61%)
Length = 981
Score = 33.5 bits (75), Expect = 0.18
Identities = 20/90 (22%), Positives = 43/90 (47%)
Frame = +2
Query: 227 ALRTVGNIVSGDDMQTQAIVNHGLLPCLLSLLTHPKKSIKKEACRTLSNITAGNREQIQA 286
ALR V +I + + GL+P ++ +L + ++ A TL + G+ E +
Sbjct: 566 ALRYVQHICQRSRSNKHVVRSAGLIPMIVDMLKSSSRKVRCRALETLRIVVEGDDENKEL 745
Query: 287 VIEAGLIAPLVNLLQNAEFDITKEVAQALT 316
+ E + +V L + +++KE +A++
Sbjct: 746 LAEGDTVRTVVKFLSH---ELSKEREEAVS 826
Score = 30.8 bits (68), Expect = 1.2
Identities = 19/89 (21%), Positives = 38/89 (42%)
Frame = +2
Query: 189 LSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGDDMQTQAIVNH 248
+ ++ + N V AG+ +V +L S AL T+ +V GDD + +
Sbjct: 578 VQHICQRSRSNKHVVRSAGLIPMIVDMLKSSSRKVRCRALETLRIVVEGDDENKELLAEG 757
Query: 249 GLLPCLLSLLTHPKKSIKKEACRTLSNIT 277
+ ++ L+H ++EA L ++
Sbjct: 758 DTVRTVVKFLSHELSKEREEAVSLLYELS 844
>TC81784 similar to PIR|T40294|T40294 hypothetical protein SPBC354.14c -
fission yeast (Schizosaccharomyces pombe), partial (30%)
Length = 1029
Score = 33.5 bits (75), Expect = 0.18
Identities = 16/50 (32%), Positives = 27/50 (54%)
Frame = +1
Query: 34 VIQSGVVPRFVQFLDKGDFPQLQLEAAWALTNIAAGTSENTKVVVDHGAV 83
+I +G + +Q L + ++Q A L N+AA + N + +VD GAV
Sbjct: 175 IIDAGFLSPLIQLLAYEENEEIQCHAISTLRNLAASSERNKRAIVDAGAV 324
>TC76366 homologue to SP|O65194|RBS_MEDSA Ribulose bisphosphate carboxylase
small chain chloroplast precursor (EC 4.1.1.39),
partial (96%)
Length = 1237
Score = 32.7 bits (73), Expect = 0.30
Identities = 18/60 (30%), Positives = 31/60 (51%)
Frame = +2
Query: 263 KSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVNLLQNAEFDITKEVAQALTNVTSGG 322
+ + K A + + +T NR + A L+AP L +A F +TK+ +T++TS G
Sbjct: 23 EGVGKMALISSAAVTTVNR------VSANLVAPFTGLKSSAGFPVTKKTNNDITSITSNG 184
>TC76307 homologue to SP|O65194|RBS_MEDSA Ribulose bisphosphate carboxylase
small chain chloroplast precursor (EC 4.1.1.39),
partial (96%)
Length = 830
Score = 32.7 bits (73), Expect = 0.30
Identities = 18/60 (30%), Positives = 31/60 (51%)
Frame = +1
Query: 263 KSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVNLLQNAEFDITKEVAQALTNVTSGG 322
+ + K A + + +T NR + A L+AP L +A F +TK+ +T++TS G
Sbjct: 76 EGVGKMALISSAAVTTVNR------VSANLVAPFTGLKSSAGFPVTKKTNNDITSITSNG 237
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.135 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,191,133
Number of Sequences: 36976
Number of extensions: 166472
Number of successful extensions: 862
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 793
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 824
length of query: 466
length of database: 9,014,727
effective HSP length: 100
effective length of query: 366
effective length of database: 5,317,127
effective search space: 1946068482
effective search space used: 1946068482
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC122160.6


